Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
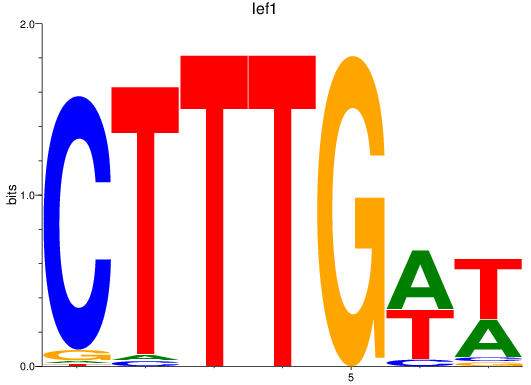
Results for lef1
Z-value: 1.40
Transcription factors associated with lef1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lef1
|
ENSDARG00000031894 | lymphoid enhancer-binding factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lef1 | dr11_v1_chr1_+_49814942_49814942 | 0.80 | 7.9e-22 | Click! |
Activity profile of lef1 motif
Sorted Z-values of lef1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_35487413 | 13.32 |
ENSDART00000102461
|
rgs8
|
regulator of G protein signaling 8 |
| chr17_+_15535501 | 9.27 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr23_+_23182037 | 9.03 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr4_-_25064510 | 8.15 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr3_-_55650771 | 7.33 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr1_+_17376922 | 7.26 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr6_-_43092175 | 6.86 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr24_+_25069609 | 6.60 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr4_-_77432218 | 6.48 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr5_-_22082918 | 6.38 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr9_-_3671911 | 6.31 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr17_-_17130942 | 6.28 |
ENSDART00000064241
|
nrxn3a
|
neurexin 3a |
| chr19_+_5291935 | 6.26 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr22_-_13851297 | 6.13 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr25_+_21829777 | 6.12 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr18_-_23875370 | 6.01 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_47582681 | 5.74 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr14_-_26177156 | 5.50 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr6_-_26559921 | 5.47 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr21_-_28920245 | 5.43 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr20_+_22666548 | 5.41 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr9_-_31278048 | 5.35 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr16_-_15988320 | 5.34 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr15_+_6109861 | 5.29 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr23_-_26077038 | 5.25 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr20_-_48485354 | 5.22 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr12_-_19865585 | 5.21 |
ENSDART00000066386
|
shisa9a
|
shisa family member 9a |
| chr3_+_29714775 | 5.13 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr8_-_34052019 | 5.12 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr5_+_60590796 | 5.10 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr20_-_40370736 | 5.08 |
ENSDART00000041229
|
fabp7b
|
fatty acid binding protein 7, brain, b |
| chr15_-_19250543 | 5.07 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr8_+_16004551 | 5.06 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr14_-_41285392 | 5.05 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr20_+_30445971 | 5.00 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr10_-_22845485 | 5.00 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr5_-_40024902 | 4.96 |
ENSDART00000017451
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr22_-_21150845 | 4.93 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr21_+_13366353 | 4.91 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr16_-_29334672 | 4.90 |
ENSDART00000162835
|
bcan
|
brevican |
| chr7_+_23875269 | 4.80 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr14_-_32403554 | 4.77 |
ENSDART00000172873
ENSDART00000173408 ENSDART00000173114 ENSDART00000185594 ENSDART00000186762 ENSDART00000010982 |
fgf13a
|
fibroblast growth factor 13a |
| chr1_+_9004719 | 4.75 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr21_+_24287403 | 4.71 |
ENSDART00000111169
|
cadm1a
|
cell adhesion molecule 1a |
| chr2_-_5135125 | 4.69 |
ENSDART00000164039
|
ptmab
|
prothymosin, alpha b |
| chr2_-_31301929 | 4.67 |
ENSDART00000191992
ENSDART00000190723 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr23_-_24450686 | 4.65 |
ENSDART00000189161
|
spen
|
spen family transcriptional repressor |
| chr4_+_12615836 | 4.62 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr6_+_40354424 | 4.58 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr25_-_4148719 | 4.57 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr23_-_10175898 | 4.54 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr6_-_35488180 | 4.53 |
ENSDART00000183258
|
rgs8
|
regulator of G protein signaling 8 |
| chr2_+_24177006 | 4.50 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr5_+_49744713 | 4.50 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr19_-_19339285 | 4.49 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr9_-_28399071 | 4.47 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr23_-_18286822 | 4.45 |
ENSDART00000136672
|
fam19a1a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1a |
| chr6_+_3828560 | 4.44 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr18_+_24922125 | 4.41 |
ENSDART00000180385
|
rgma
|
repulsive guidance molecule family member a |
| chr10_+_21780250 | 4.41 |
ENSDART00000183782
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr2_-_30784198 | 4.36 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr21_-_27010796 | 4.35 |
ENSDART00000065398
ENSDART00000144342 ENSDART00000126542 |
ppp1r14ba
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ba |
| chr2_+_29976419 | 4.34 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr9_+_42066030 | 4.30 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr10_-_27049170 | 4.30 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr8_-_17064243 | 4.29 |
ENSDART00000185313
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr5_-_35301800 | 4.27 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr25_-_11057753 | 4.25 |
ENSDART00000186551
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr6_+_29861288 | 4.22 |
ENSDART00000166782
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr7_-_38612230 | 4.22 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr25_-_32869794 | 4.17 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr14_+_22172047 | 4.15 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr6_+_46341306 | 4.14 |
ENSDART00000111905
|
BX649498.1
|
|
| chr2_-_30460293 | 4.12 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr17_-_15657029 | 4.11 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr12_+_7497882 | 4.00 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr17_+_24632440 | 3.88 |
ENSDART00000157092
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr2_-_16562505 | 3.86 |
ENSDART00000156406
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr19_+_21408709 | 3.84 |
ENSDART00000186194
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr18_-_7137153 | 3.81 |
ENSDART00000019571
|
cd9a
|
CD9 molecule a |
| chr20_-_37522569 | 3.80 |
ENSDART00000177011
ENSDART00000058502 |
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr7_-_28463106 | 3.79 |
ENSDART00000137799
|
trim66
|
tripartite motif containing 66 |
| chr14_-_21618005 | 3.78 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr7_-_56793739 | 3.77 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr7_+_27977065 | 3.77 |
ENSDART00000089574
|
tub
|
tubby bipartite transcription factor |
| chr21_-_25604603 | 3.70 |
ENSDART00000133134
ENSDART00000139460 |
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr7_+_57795974 | 3.70 |
ENSDART00000148369
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr5_-_30418636 | 3.68 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr8_-_34051548 | 3.65 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr21_+_13387965 | 3.64 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr8_-_7504146 | 3.64 |
ENSDART00000148339
ENSDART00000140098 |
cdk20
|
cyclin-dependent kinase 20 |
| chr7_+_57836841 | 3.64 |
ENSDART00000136175
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr22_-_16042243 | 3.61 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr17_+_52822422 | 3.57 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr14_+_8127893 | 3.55 |
ENSDART00000169091
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr5_-_33460959 | 3.54 |
ENSDART00000085636
|
si:ch211-182d3.1
|
si:ch211-182d3.1 |
| chr16_-_37964325 | 3.54 |
ENSDART00000148801
|
mrap2a
|
melanocortin 2 receptor accessory protein 2a |
| chr12_-_31103187 | 3.54 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr25_-_12788370 | 3.53 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr15_+_7992906 | 3.53 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr23_-_18982499 | 3.52 |
ENSDART00000012507
|
bcl2l1
|
bcl2-like 1 |
| chr20_-_23219964 | 3.52 |
ENSDART00000144933
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr13_+_30506781 | 3.49 |
ENSDART00000110884
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr13_+_4405282 | 3.46 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr22_+_20208185 | 3.46 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr9_+_29585943 | 3.46 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr10_+_21650828 | 3.43 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr23_-_35694171 | 3.43 |
ENSDART00000077539
|
tuba1c
|
tubulin, alpha 1c |
| chr6_+_23810529 | 3.42 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr1_+_49814461 | 3.42 |
ENSDART00000132405
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr23_-_36449111 | 3.40 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr10_+_21783213 | 3.38 |
ENSDART00000168899
|
pcdh1g33
|
protocadherin 1 gamma 33 |
| chr7_-_34329527 | 3.38 |
ENSDART00000173454
|
madd
|
MAP-kinase activating death domain |
| chr20_+_31185881 | 3.37 |
ENSDART00000153391
|
otofa
|
otoferlin a |
| chr16_-_22775480 | 3.36 |
ENSDART00000141778
ENSDART00000145585 ENSDART00000125963 ENSDART00000127570 |
pbxip1b
|
pre-B-cell leukemia homeobox interacting protein 1b |
| chr5_+_24156170 | 3.36 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr11_+_36989696 | 3.33 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr5_-_26093945 | 3.32 |
ENSDART00000010199
ENSDART00000145096 |
fam219ab
|
family with sequence similarity 219, member Ab |
| chr18_-_38088099 | 3.31 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr17_-_20897250 | 3.31 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr11_+_26604224 | 3.31 |
ENSDART00000030453
ENSDART00000168895 ENSDART00000159505 |
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr9_+_38168012 | 3.31 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr25_+_18953575 | 3.31 |
ENSDART00000155610
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr17_+_9308425 | 3.31 |
ENSDART00000188283
ENSDART00000183311 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr4_-_27117112 | 3.29 |
ENSDART00000034534
|
zbed4
|
zinc finger, BED-type containing 4 |
| chr9_+_6009077 | 3.26 |
ENSDART00000057484
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr6_+_296130 | 3.22 |
ENSDART00000073985
|
rbfox2
|
RNA binding fox-1 homolog 2 |
| chr1_-_26131088 | 3.22 |
ENSDART00000193973
ENSDART00000054209 |
cdkn2a/b
|
cyclin-dependent kinase inhibitor 2A/B (p15, inhibits CDK4) |
| chr16_+_25011994 | 3.19 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr2_+_33368414 | 3.19 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr24_-_6644734 | 3.17 |
ENSDART00000167391
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr8_+_16407884 | 3.15 |
ENSDART00000133742
|
cdkn2c
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr5_+_20823409 | 3.14 |
ENSDART00000093185
ENSDART00000142894 |
limk2
|
LIM domain kinase 2 |
| chr14_+_41406321 | 3.13 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr19_-_13774502 | 3.13 |
ENSDART00000159711
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr20_-_9436521 | 3.13 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr13_+_38430466 | 3.12 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr13_-_31389661 | 3.11 |
ENSDART00000134630
|
zdhhc16a
|
zinc finger, DHHC-type containing 16a |
| chr15_-_23761580 | 3.11 |
ENSDART00000137918
|
bbc3
|
BCL2 binding component 3 |
| chr15_+_32821392 | 3.11 |
ENSDART00000158272
|
dclk1b
|
doublecortin-like kinase 1b |
| chr5_-_24231139 | 3.10 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr9_-_35155089 | 3.10 |
ENSDART00000077901
|
appb
|
amyloid beta (A4) precursor protein b |
| chr7_+_20965880 | 3.10 |
ENSDART00000062003
|
efnb3b
|
ephrin-B3b |
| chr2_-_8017579 | 3.10 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr14_+_11762991 | 3.08 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr19_-_30510259 | 3.08 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr21_+_23953181 | 3.08 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr22_-_21845685 | 3.07 |
ENSDART00000105564
|
aes
|
amino-terminal enhancer of split |
| chr2_+_25658112 | 3.06 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr7_-_48805181 | 3.05 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr24_+_5840258 | 3.04 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr6_+_6780873 | 3.03 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr22_+_8753092 | 3.01 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr1_+_44173245 | 3.01 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr14_+_33525196 | 3.01 |
ENSDART00000085335
|
zdhhc9
|
zinc finger, DHHC-type containing 9 |
| chr9_+_25568839 | 3.00 |
ENSDART00000177342
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr22_+_28803739 | 2.98 |
ENSDART00000129476
ENSDART00000189726 |
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr23_-_12015139 | 2.98 |
ENSDART00000110627
ENSDART00000193988 ENSDART00000184528 |
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr4_-_4261673 | 2.98 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr13_+_28732101 | 2.94 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr7_-_9803154 | 2.94 |
ENSDART00000055593
|
aldh1a3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr2_+_25657958 | 2.93 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr14_+_6963312 | 2.92 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr3_+_23248704 | 2.92 |
ENSDART00000156032
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr24_-_32408404 | 2.92 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr21_+_45496442 | 2.91 |
ENSDART00000164880
|
si:dkey-223p19.1
|
si:dkey-223p19.1 |
| chr14_-_2270973 | 2.90 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr22_+_11972786 | 2.87 |
ENSDART00000105788
|
mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr12_+_25640480 | 2.87 |
ENSDART00000105608
|
prkcea
|
protein kinase C, epsilon a |
| chr24_-_28259127 | 2.87 |
ENSDART00000149589
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr17_+_36588281 | 2.87 |
ENSDART00000076115
|
htr1b
|
5-hydroxytryptamine (serotonin) receptor 1B |
| chr11_-_3865472 | 2.86 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr14_-_1990290 | 2.85 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr3_-_30861177 | 2.84 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr7_+_14632157 | 2.83 |
ENSDART00000161264
|
ntrk3b
|
neurotrophic tyrosine kinase, receptor, type 3b |
| chr2_-_49031303 | 2.81 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr3_-_55650417 | 2.81 |
ENSDART00000171441
|
axin2
|
axin 2 (conductin, axil) |
| chr5_+_67662430 | 2.80 |
ENSDART00000137700
ENSDART00000142586 |
si:dkey-70b23.2
|
si:dkey-70b23.2 |
| chr2_+_24177190 | 2.80 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr17_+_47174525 | 2.79 |
ENSDART00000156831
|
CLIP4
|
si:dkeyp-47f9.4 |
| chr24_+_21514283 | 2.79 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr12_+_41510492 | 2.79 |
ENSDART00000170976
ENSDART00000176164 |
kif5bb
|
kinesin family member 5B, b |
| chr6_-_30859656 | 2.78 |
ENSDART00000156235
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr23_-_24682244 | 2.76 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr18_-_23875219 | 2.76 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr21_-_25722834 | 2.76 |
ENSDART00000101208
|
abhd11
|
abhydrolase domain containing 11 |
| chr5_-_46896541 | 2.76 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr23_+_21638258 | 2.75 |
ENSDART00000104188
|
igsf21b
|
immunoglobin superfamily, member 21b |
| chr9_-_44642108 | 2.75 |
ENSDART00000086202
|
pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_+_44713135 | 2.75 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr20_+_474288 | 2.75 |
ENSDART00000026794
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr23_+_21544227 | 2.74 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr14_+_45306073 | 2.74 |
ENSDART00000034606
ENSDART00000173301 |
sfxn5b
|
sideroflexin 5b |
| chr2_+_27010439 | 2.72 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr23_+_20523617 | 2.72 |
ENSDART00000176404
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr8_-_1264893 | 2.71 |
ENSDART00000190371
|
cdc14b
|
cell division cycle 14B |
| chr16_+_32136550 | 2.71 |
ENSDART00000147526
|
sphk2
|
sphingosine kinase 2 |
| chr16_-_31451720 | 2.69 |
ENSDART00000146886
|
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr2_+_42135719 | 2.67 |
ENSDART00000008268
|
cyp7b1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr8_+_32516160 | 2.67 |
ENSDART00000061786
|
ncs1b
|
neuronal calcium sensor 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of lef1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 17.8 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 2.8 | 8.3 | GO:0060843 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 1.5 | 10.5 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 1.5 | 4.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.2 | 3.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 1.2 | 3.5 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 1.1 | 3.4 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 1.1 | 5.5 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 1.1 | 7.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 1.0 | 3.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 1.0 | 10.3 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 1.0 | 1.0 | GO:2000742 | regulation of anterior head development(GO:2000742) |
| 1.0 | 2.9 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.9 | 12.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.9 | 3.5 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.9 | 6.9 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.8 | 2.4 | GO:1903792 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.8 | 3.2 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.8 | 2.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.8 | 9.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.8 | 5.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.7 | 2.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.7 | 2.0 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.7 | 2.7 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.7 | 3.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.7 | 2.0 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.6 | 5.7 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.6 | 3.8 | GO:0022029 | telencephalon cell migration(GO:0022029) |
| 0.6 | 4.4 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.6 | 3.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.6 | 3.1 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.6 | 1.9 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.6 | 14.3 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.6 | 5.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.6 | 3.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.6 | 2.3 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.6 | 2.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.6 | 2.2 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.6 | 4.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.6 | 5.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.5 | 7.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 2.7 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.5 | 1.6 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.5 | 4.5 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.5 | 2.5 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.5 | 2.0 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.5 | 1.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.5 | 4.8 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.5 | 3.4 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 | 1.9 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.5 | 1.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.5 | 6.1 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.5 | 6.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.5 | 1.4 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.5 | 6.0 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.5 | 3.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.5 | 2.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.4 | 4.5 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.4 | 2.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.4 | 1.3 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.4 | 1.7 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.4 | 3.0 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.4 | 2.0 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.4 | 10.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.4 | 2.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.4 | 5.5 | GO:0001840 | neural plate development(GO:0001840) |
| 0.4 | 1.4 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.3 | 12.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 1.4 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.3 | 3.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 5.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.3 | 2.3 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.3 | 4.2 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.3 | 1.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.3 | 6.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 3.5 | GO:0097009 | protein localization to cell surface(GO:0034394) energy homeostasis(GO:0097009) |
| 0.3 | 1.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.3 | 4.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.3 | 3.8 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.3 | 1.2 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.3 | 2.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.3 | 2.4 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.3 | 2.9 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 2.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.3 | 1.4 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.3 | 4.2 | GO:0001964 | startle response(GO:0001964) |
| 0.3 | 3.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.3 | 1.1 | GO:1901827 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.3 | 0.3 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.3 | 1.9 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.3 | 4.4 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.3 | 0.8 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.3 | 3.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.3 | 4.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.3 | 1.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.3 | 0.8 | GO:1903011 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.3 | 1.6 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.3 | 1.3 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.3 | 2.9 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.3 | 4.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 2.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 | 2.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.3 | 2.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.3 | 2.6 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.3 | 2.1 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.3 | 2.6 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.3 | 4.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.3 | 8.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 1.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 1.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.2 | 2.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 4.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 0.7 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.2 | 3.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 7.3 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.2 | 3.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 0.9 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 0.7 | GO:0002370 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of mast cell activation(GO:0033005) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.2 | 1.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 2.0 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.2 | 2.2 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.2 | 3.9 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 2.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 1.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.2 | 6.2 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.2 | 3.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 4.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 0.4 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.2 | 2.6 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.2 | 2.6 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.2 | 0.6 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 1.0 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.2 | 1.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 0.8 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.2 | 2.9 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.2 | 0.6 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 1.5 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.2 | 4.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 1.8 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 3.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 3.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 2.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.2 | 2.4 | GO:1901797 | negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.2 | 3.4 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.2 | 2.2 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.2 | 1.7 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.2 | 4.1 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.2 | 1.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 0.6 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.2 | 1.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 0.9 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 4.0 | GO:0021761 | limbic system development(GO:0021761) |
| 0.2 | 1.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 1.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 8.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.2 | 2.1 | GO:0010575 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.9 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 7.0 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.1 | 5.0 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 2.1 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 3.5 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 2.6 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 1.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 4.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 2.5 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 4.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 3.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.4 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 4.4 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.1 | 2.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 1.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 7.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 3.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 1.5 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 3.8 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 3.7 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.1 | 24.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 3.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 2.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 2.9 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 3.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 8.1 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.1 | 8.6 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 | 3.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 10.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 2.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.0 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.3 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.5 | GO:0044819 | mitotic G1/S transition checkpoint(GO:0044819) |
| 0.1 | 0.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.8 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 0.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 2.9 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 2.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 1.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 1.2 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.2 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 0.5 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.1 | 0.8 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 2.0 | GO:0050890 | cognition(GO:0050890) |
| 0.1 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.9 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.1 | 3.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 3.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 1.0 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 5.8 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.1 | 5.4 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 1.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.0 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 3.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.7 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 3.7 | GO:0018210 | peptidyl-threonine modification(GO:0018210) |
| 0.1 | 1.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 1.9 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 1.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 1.3 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 1.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 10.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 1.6 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 2.5 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.1 | 1.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 4.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.6 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.1 | 2.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 6.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.6 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.1 | 1.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 1.6 | GO:0035138 | pectoral fin morphogenesis(GO:0035138) |
| 0.1 | 0.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.7 | GO:0030534 | adult behavior(GO:0030534) |
| 0.1 | 0.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 4.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.9 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.5 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.9 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 2.2 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 1.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 1.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 6.7 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.4 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) reticulophagy(GO:0061709) single-organism membrane invagination(GO:1902534) |
| 0.0 | 1.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 5.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 2.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.4 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.0 | 0.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.7 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 5.4 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.2 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.0 | 0.4 | GO:0098868 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.0 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 6.4 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 1.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.3 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 1.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0007308 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 1.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 13.4 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.4 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 1.2 | GO:0050769 | positive regulation of neurogenesis(GO:0050769) |
| 0.0 | 0.2 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 2.0 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 1.9 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 1.3 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.7 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 2.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.9 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.8 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.2 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) spinal cord radial glial cell differentiation(GO:0021531) optic placode formation involved in camera-type eye formation(GO:0046619) radial glial cell differentiation(GO:0060019) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.5 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.0 | 0.6 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 0.3 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 1.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 1.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.7 | GO:0030902 | hindbrain development(GO:0030902) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0034372 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) high-density lipoprotein particle assembly(GO:0034380) reverse cholesterol transport(GO:0043691) |
| 0.0 | 3.7 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.0 | 0.7 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) positive regulation of cysteine-type endopeptidase activity(GO:2001056) |
| 0.0 | 0.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 3.5 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.2 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 1.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.8 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.9 | 3.5 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.9 | 15.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.7 | 2.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.5 | 1.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.5 | 3.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 3.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.4 | 4.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 5.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.4 | 3.9 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 3.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 2.0 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.3 | 13.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 8.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 5.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 3.8 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.3 | 1.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.3 | 10.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 0.8 | GO:0072380 | TRC complex(GO:0072380) |
| 0.3 | 7.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 2.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 2.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 2.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 0.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 0.9 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 10.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 1.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 2.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 2.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 3.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 4.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 2.5 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.2 | 3.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 3.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 9.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 7.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 1.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 9.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 9.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 3.0 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 5.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 5.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 8.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 2.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 2.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.7 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.9 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 2.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 2.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 5.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 5.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 3.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 4.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 2.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 1.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 4.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 1.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 1.1 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 5.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 2.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 4.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.4 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 9.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 7.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 1.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 12.1 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.1 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 2.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 17.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.8 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 4.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 7.6 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 4.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 14.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 6.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 27.0 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 4.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 5.0 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 86.7 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.2 | GO:0032993 | nucleosome(GO:0000786) protein-DNA complex(GO:0032993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.6 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 1.8 | 5.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 1.6 | 6.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.5 | 6.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.5 | 4.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.2 | 5.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 1.1 | 8.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 1.0 | 9.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 1.0 | 3.0 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.9 | 2.7 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.9 | 3.5 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.8 | 4.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.8 | 7.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.7 | 5.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.7 | 2.0 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.6 | 3.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.6 | 4.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.6 | 1.7 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.6 | 3.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.5 | 1.6 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.5 | 7.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.5 | 1.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 3.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.5 | 2.0 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.5 | 2.8 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 6.1 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.5 | 2.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.5 | 6.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 2.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 2.6 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.4 | 2.5 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.4 | 1.2 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.4 | 3.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.4 | 8.3 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.4 | 2.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.4 | 3.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.4 | 3.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.4 | 1.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 2.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 3.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 1.4 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.3 | 1.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 4.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 2.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 1.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 5.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 2.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 4.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 1.9 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.3 | 1.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 1.6 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.3 | 2.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.3 | 2.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 2.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 1.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.3 | 1.2 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.3 | 2.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 1.5 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.3 | 1.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 1.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 7.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.3 | 3.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 1.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.3 | 4.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 3.0 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.3 | 3.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.3 | 4.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.3 | 6.6 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 7.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 1.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 1.7 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 4.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 1.8 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.9 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.2 | 1.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 2.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 8.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 2.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 4.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 1.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 3.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 2.8 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 9.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 3.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 9.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 1.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 0.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.9 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 2.5 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 3.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 2.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 1.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 7.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 3.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 6.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 1.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 7.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 1.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.7 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 1.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 3.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 1.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 3.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 3.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 4.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 3.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.6 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 2.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 6.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.9 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 4.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.4 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 1.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 1.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.7 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 2.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 3.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.8 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 2.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.8 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 2.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.7 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 2.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 2.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 6.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.9 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 7.1 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 2.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 7.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 34.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.3 | GO:0051903 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.1 | 1.3 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 1.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 7.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 10.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 2.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 8.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 1.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 2.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 2.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.8 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 2.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 8.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.6 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 2.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 9.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.6 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.1 | 0.4 | GO:0035255 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 4.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 4.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 3.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.0 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 4.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.7 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 12.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.0 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 11.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 7.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 7.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 11.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 24.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 2.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 1.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.2 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 1.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 7.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 2.4 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 25.3 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 9.0 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 13.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.4 | 3.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 6.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.4 | 8.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.3 | 8.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 4.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 8.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 1.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 4.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 8.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 2.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 4.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 2.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 7.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 2.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 4.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 7.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 3.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 7.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 6.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.2 | 5.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 5.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 2.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.7 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.7 | 6.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.5 | 3.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.5 | 2.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 6.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 5.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 8.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.4 | 2.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.4 | 1.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 1.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.3 | 6.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 3.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 2.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 7.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.3 | 7.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 4.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 2.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 4.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 3.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 5.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 27.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 2.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 1.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 11.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 7.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 1.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 2.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 1.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 1.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.2 | 2.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 3.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 4.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 5.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.1 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 8.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 3.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 4.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 5.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |