Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
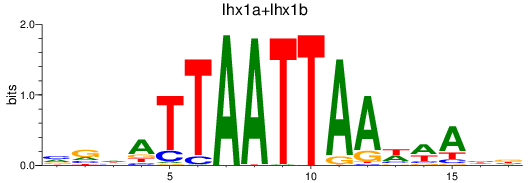
Results for lhx1a+lhx1b
Z-value: 0.40
Transcription factors associated with lhx1a+lhx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx1b
|
ENSDARG00000007944 | LIM homeobox 1b |
|
lhx1a
|
ENSDARG00000014018 | LIM homeobox 1a |
|
lhx1b
|
ENSDARG00000111635 | LIM homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx1b | dr11_v1_chr5_+_56268436_56268436 | 0.21 | 4.5e-02 | Click! |
| lhx1a | dr11_v1_chr15_-_27710513_27710575 | 0.16 | 1.2e-01 | Click! |
Activity profile of lhx1a+lhx1b motif
Sorted Z-values of lhx1a+lhx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_32818607 | 5.47 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr9_+_34641237 | 3.39 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr24_-_26328721 | 3.38 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr4_+_9669717 | 2.44 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr25_+_4751879 | 2.42 |
ENSDART00000169465
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr24_+_2495197 | 2.24 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr6_-_30839763 | 2.21 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr5_+_4054704 | 2.17 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr3_-_19368435 | 1.94 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr7_-_38658411 | 1.84 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr19_+_2631565 | 1.70 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr18_-_17485419 | 1.66 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr23_+_4689626 | 1.55 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr6_-_40029423 | 1.55 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr15_-_43284021 | 1.50 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr10_-_11261565 | 1.41 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr5_+_60590796 | 1.38 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr12_-_17712393 | 1.37 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr21_-_35419486 | 1.31 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr1_+_6172786 | 1.27 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr9_-_9415000 | 1.26 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr17_-_16422654 | 1.23 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr17_-_45040813 | 1.18 |
ENSDART00000075514
|
entpd5a
|
ectonucleoside triphosphate diphosphohydrolase 5a |
| chr20_-_45812144 | 1.18 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr2_+_20605925 | 1.11 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr16_-_28856112 | 1.07 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr5_+_61301525 | 1.03 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr3_+_39853788 | 1.03 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr21_-_20939488 | 1.02 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr19_+_46158078 | 0.98 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr9_+_24159280 | 0.98 |
ENSDART00000184624
ENSDART00000178422 |
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr3_-_15999501 | 0.97 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr6_-_50730749 | 0.96 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr6_-_11768198 | 0.93 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr1_+_10318089 | 0.90 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr11_-_40204621 | 0.88 |
ENSDART00000086287
|
znf362a
|
zinc finger protein 362a |
| chr24_-_35767501 | 0.88 |
ENSDART00000105680
ENSDART00000042290 ENSDART00000166264 |
dtna
|
dystrobrevin, alpha |
| chr19_+_43297546 | 0.86 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr19_-_5769728 | 0.83 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr6_+_7444899 | 0.82 |
ENSDART00000053775
|
arf3b
|
ADP-ribosylation factor 3b |
| chr4_+_16885854 | 0.78 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr2_-_16217344 | 0.76 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr12_+_7497882 | 0.76 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr16_+_2820340 | 0.75 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr9_-_14504834 | 0.71 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr9_-_47472998 | 0.70 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr19_-_5769553 | 0.69 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr16_+_54209504 | 0.69 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr22_-_28226948 | 0.68 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr15_-_20939579 | 0.66 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr6_+_35052721 | 0.66 |
ENSDART00000191090
ENSDART00000082940 |
uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr13_-_29420885 | 0.65 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr12_+_45200744 | 0.64 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr6_-_44402358 | 0.62 |
ENSDART00000193007
ENSDART00000193603 |
pdzrn3b
|
PDZ domain containing RING finger 3b |
| chr3_-_26806032 | 0.62 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr1_-_51038885 | 0.61 |
ENSDART00000035150
|
spast
|
spastin |
| chr13_+_11440389 | 0.60 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr10_-_11261386 | 0.58 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr11_-_25733910 | 0.57 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr6_+_11989537 | 0.55 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr4_+_12966640 | 0.54 |
ENSDART00000113357
|
vhll
|
von Hippel-Lindau tumor suppressor like |
| chr17_+_45405821 | 0.54 |
ENSDART00000189482
ENSDART00000177422 |
tagapa
|
T cell activation RhoGTPase activating protein a |
| chr24_+_28953089 | 0.54 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr13_-_29421331 | 0.53 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr15_-_17618800 | 0.53 |
ENSDART00000157185
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr1_+_51039558 | 0.52 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr1_-_669717 | 0.50 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr16_-_12173554 | 0.49 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr10_+_17235370 | 0.49 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr3_-_56541723 | 0.49 |
ENSDART00000156398
ENSDART00000050576 ENSDART00000184874 |
si:ch211-189a21.1
cyth1a
|
si:ch211-189a21.1 cytohesin 1a |
| chr22_-_12160283 | 0.48 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr22_-_8725768 | 0.48 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr25_-_25058508 | 0.48 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr7_-_17297156 | 0.48 |
ENSDART00000161336
|
nitr11a
|
novel immune-type receptor 11a |
| chr11_+_30663300 | 0.47 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr4_+_52212057 | 0.44 |
ENSDART00000165627
|
znf1034
|
zinc finger protein 1034 |
| chr3_+_26342768 | 0.43 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr23_-_31969786 | 0.43 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr10_+_45089820 | 0.43 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr21_-_2415808 | 0.42 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr22_-_10156581 | 0.39 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr25_+_18711804 | 0.39 |
ENSDART00000011149
|
fam185a
|
family with sequence similarity 185, member A |
| chr3_-_11624694 | 0.36 |
ENSDART00000127157
|
hlfa
|
hepatic leukemia factor a |
| chr15_-_22074315 | 0.35 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr12_+_48480632 | 0.35 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr4_+_72723304 | 0.35 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr14_-_413273 | 0.34 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr3_-_26787430 | 0.34 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr9_+_24159725 | 0.32 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr5_-_31901468 | 0.32 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr3_+_36424055 | 0.32 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr7_+_20471315 | 0.31 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr21_+_25802190 | 0.30 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr4_+_77943184 | 0.30 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr15_-_17619306 | 0.29 |
ENSDART00000184011
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr8_-_16712111 | 0.28 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr6_-_34838397 | 0.27 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr22_-_19552796 | 0.27 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr23_-_16485190 | 0.25 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr6_-_35052145 | 0.25 |
ENSDART00000073970
ENSDART00000185790 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr16_-_12173399 | 0.24 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr1_-_15797663 | 0.24 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr1_+_54124209 | 0.23 |
ENSDART00000187730
|
LO017722.1
|
|
| chr21_-_17482465 | 0.22 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr4_+_57881965 | 0.22 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr4_-_2219705 | 0.21 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr6_-_21582444 | 0.20 |
ENSDART00000151339
|
si:dkey-43k4.3
|
si:dkey-43k4.3 |
| chr8_+_50953776 | 0.20 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr8_-_53926228 | 0.20 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr23_-_4409668 | 0.19 |
ENSDART00000081823
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr6_-_35052388 | 0.19 |
ENSDART00000181000
ENSDART00000170116 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr5_-_23843636 | 0.19 |
ENSDART00000193280
|
GBGT1 (1 of many)
|
si:ch211-135f11.5 |
| chr22_-_22242884 | 0.19 |
ENSDART00000020937
|
hdgfl2
|
HDGF like 2 |
| chr15_-_14552101 | 0.19 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr5_+_41477954 | 0.18 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr12_-_48006835 | 0.17 |
ENSDART00000108989
|
adamts14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr24_-_40860603 | 0.17 |
ENSDART00000188032
|
CU633479.7
|
|
| chr25_-_13490744 | 0.16 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr9_+_37152564 | 0.16 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr5_-_38094130 | 0.16 |
ENSDART00000131831
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr23_-_17509656 | 0.15 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr10_+_42423318 | 0.14 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr25_+_4855549 | 0.12 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr10_-_2524917 | 0.12 |
ENSDART00000188642
|
CU856539.1
|
|
| chr17_+_31914877 | 0.11 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr10_-_2524297 | 0.11 |
ENSDART00000192475
|
CU856539.1
|
|
| chr18_+_46382484 | 0.11 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr20_+_11731039 | 0.10 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr17_+_22577472 | 0.10 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr4_+_5255041 | 0.10 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr22_-_13466246 | 0.10 |
ENSDART00000134035
|
cntnap5b
|
contactin associated protein-like 5b |
| chr5_+_41477526 | 0.08 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr3_+_45687266 | 0.08 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr17_-_40956035 | 0.08 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr21_-_22635245 | 0.07 |
ENSDART00000115224
ENSDART00000101782 |
nectin1a
|
nectin cell adhesion molecule 1a |
| chr3_-_48716422 | 0.07 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr5_-_30620625 | 0.07 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr24_+_26329018 | 0.07 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr20_-_28642061 | 0.06 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr17_+_49281597 | 0.06 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr23_-_1660708 | 0.06 |
ENSDART00000175138
|
CU693481.1
|
|
| chr24_+_26328787 | 0.05 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr17_+_28533102 | 0.04 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr16_+_13965923 | 0.02 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr23_+_17509794 | 0.01 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx1a+lhx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.4 | 1.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.6 | GO:0034214 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.2 | 1.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 2.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 1.4 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 1.2 | GO:0072506 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 | 0.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 3.4 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 1.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 1.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.7 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.1 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.3 | GO:0016122 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 2.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.2 | GO:0071498 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.7 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 1.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 1.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 5.5 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.1 | 1.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.2 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 1.2 | GO:0030814 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 1.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 1.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0044406 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 1.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.6 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 1.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.0 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 1.8 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 0.7 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.1 | 0.5 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.3 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 2.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 1.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 1.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.9 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 2.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.3 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 1.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |