Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
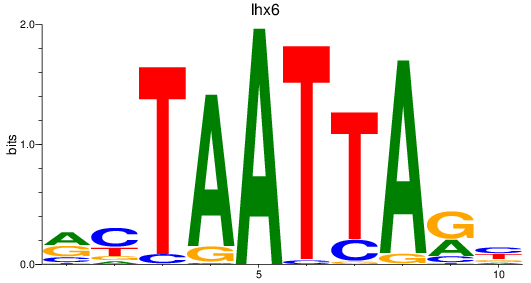
Results for lhx6
Z-value: 0.76
Transcription factors associated with lhx6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx6
|
ENSDARG00000006896 | LIM homeobox 6 |
|
lhx6
|
ENSDARG00000112520 | LIM homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx6 | dr11_v1_chr10_+_9372702_9372702 | -0.10 | 3.2e-01 | Click! |
Activity profile of lhx6 motif
Sorted Z-values of lhx6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_15433671 | 11.40 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433518 | 11.34 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr20_-_9436521 | 11.01 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr5_+_64732036 | 8.22 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr4_+_21129752 | 8.18 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr5_+_64732270 | 7.82 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr7_+_30787903 | 6.93 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr1_-_50859053 | 6.46 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr8_+_16025554 | 6.04 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr5_-_50992690 | 5.56 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr16_+_46111849 | 5.15 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr14_+_14662116 | 5.10 |
ENSDART00000161693
|
cetn2
|
centrin, EF-hand protein, 2 |
| chr25_-_13842618 | 5.05 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr7_-_49594995 | 4.99 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr24_-_21923930 | 4.98 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr5_+_61301525 | 4.68 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr2_+_50608099 | 4.59 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr14_-_33872092 | 4.57 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr4_+_4849789 | 4.42 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr1_+_25801648 | 4.26 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr13_+_15838151 | 4.21 |
ENSDART00000008987
|
klc1a
|
kinesin light chain 1a |
| chr10_-_27049170 | 4.21 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr21_+_15824182 | 4.14 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr17_-_36896560 | 4.09 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr19_-_9882821 | 4.08 |
ENSDART00000147128
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr19_-_5103313 | 4.00 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr14_-_33872616 | 3.91 |
ENSDART00000162840
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr2_+_26237322 | 3.86 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr21_-_39177564 | 3.76 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr4_+_11384891 | 3.69 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr19_-_5103141 | 3.66 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr3_+_45365098 | 3.63 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr11_-_44801968 | 3.60 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr3_+_45364849 | 3.54 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr8_+_49778486 | 3.53 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr16_+_47207691 | 3.49 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr13_-_31435137 | 3.48 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr5_+_22098591 | 3.44 |
ENSDART00000143676
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr4_-_17629444 | 3.43 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr13_-_29420885 | 3.41 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr2_+_20332044 | 3.35 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr7_+_56577522 | 3.34 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr21_+_28958471 | 3.25 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr13_+_38521152 | 3.15 |
ENSDART00000145292
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr9_+_38372216 | 3.14 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr9_+_17983463 | 3.14 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr17_+_24722646 | 3.14 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr9_+_30108641 | 2.99 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr20_-_29864390 | 2.98 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr4_-_1801519 | 2.97 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr19_-_30524952 | 2.96 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr4_-_28158335 | 2.95 |
ENSDART00000134605
|
gramd4a
|
GRAM domain containing 4a |
| chr14_+_8940326 | 2.93 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr7_+_56577906 | 2.92 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr22_+_27090136 | 2.90 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr24_+_16547035 | 2.90 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr1_-_18811517 | 2.90 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr3_+_34919810 | 2.89 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr19_+_233143 | 2.88 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr16_+_34531486 | 2.78 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr20_-_23226453 | 2.75 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr10_+_21726853 | 2.74 |
ENSDART00000162869
|
pcdh1g14
|
protocadherin 1 gamma 14 |
| chr11_+_11267829 | 2.72 |
ENSDART00000026814
ENSDART00000173346 ENSDART00000151926 |
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr13_-_24311628 | 2.67 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr15_-_9272328 | 2.67 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr20_-_46362606 | 2.62 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr21_+_26748141 | 2.61 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr18_+_8320165 | 2.61 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr7_-_28148310 | 2.57 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr23_+_40460333 | 2.56 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr11_+_18873619 | 2.54 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr5_+_36768674 | 2.51 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr9_-_20372977 | 2.49 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr22_+_5176255 | 2.48 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr20_-_14114078 | 2.47 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr6_-_6487876 | 2.47 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr2_+_6253246 | 2.46 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr23_+_28582865 | 2.43 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr2_+_40294313 | 2.42 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr15_-_33925851 | 2.42 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr11_+_38280454 | 2.41 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr13_+_19322686 | 2.37 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr5_+_60590796 | 2.27 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr23_-_10786400 | 2.27 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr18_+_7639401 | 2.26 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr22_-_20166660 | 2.25 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr10_-_5581487 | 2.25 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr12_-_4781801 | 2.24 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr11_+_18873113 | 2.23 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr23_-_18130264 | 2.22 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr18_-_2433011 | 2.21 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr20_-_48485354 | 2.15 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr17_-_49438873 | 2.13 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr6_-_55585423 | 2.12 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr5_+_29851433 | 2.10 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr5_+_37903790 | 2.09 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr21_+_42226113 | 2.07 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr15_+_23799461 | 2.04 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr22_-_24818066 | 2.01 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr6_+_21001264 | 1.95 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr7_-_30174882 | 1.92 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr2_-_32558795 | 1.87 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr6_+_39232245 | 1.83 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr1_+_33668236 | 1.79 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr11_-_42554290 | 1.77 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr5_-_16996482 | 1.77 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr1_+_18811679 | 1.77 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr25_-_19224298 | 1.77 |
ENSDART00000149917
|
acanb
|
aggrecan b |
| chr4_+_2655358 | 1.76 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr18_+_17827149 | 1.76 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr17_+_49281597 | 1.76 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr8_+_49778756 | 1.74 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr1_-_12109216 | 1.73 |
ENSDART00000079930
|
mttp
|
microsomal triglyceride transfer protein |
| chr1_-_44701313 | 1.72 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr24_-_25004553 | 1.64 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr20_+_41021054 | 1.63 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr11_+_28476298 | 1.62 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr19_-_42588510 | 1.61 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr3_-_46817499 | 1.61 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr20_+_29209615 | 1.58 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr3_+_62353650 | 1.56 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr5_-_41307550 | 1.56 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr4_-_73756673 | 1.56 |
ENSDART00000174274
ENSDART00000192913 ENSDART00000113546 |
BX855614.4
si:dkey-262g12.14
zgc:171551
|
si:dkey-262g12.14 zgc:171551 |
| chr12_+_41697664 | 1.55 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr20_+_29209767 | 1.54 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr5_-_64831207 | 1.52 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr17_-_40956035 | 1.51 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr6_+_28208973 | 1.51 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr7_-_59159253 | 1.50 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr15_+_17345609 | 1.50 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr7_-_35708450 | 1.48 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr14_-_16082806 | 1.46 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr8_-_49766205 | 1.46 |
ENSDART00000137941
ENSDART00000097919 ENSDART00000147309 |
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr21_-_14175838 | 1.44 |
ENSDART00000111659
|
whrna
|
whirlin a |
| chr3_-_43356082 | 1.42 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr5_-_19006290 | 1.41 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr17_-_200316 | 1.40 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr15_-_18115540 | 1.40 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr6_+_45918981 | 1.40 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr15_+_6114109 | 1.40 |
ENSDART00000184937
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr11_-_40728380 | 1.39 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr11_+_41540862 | 1.38 |
ENSDART00000173210
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr2_+_57848844 | 1.37 |
ENSDART00000037279
|
plekhj1
|
pleckstrin homology domain containing, family J member 1 |
| chr24_-_39518599 | 1.37 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr16_-_26296477 | 1.36 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr22_-_21897203 | 1.36 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr7_+_38811800 | 1.36 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr10_-_34002185 | 1.34 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr2_+_35728033 | 1.33 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr7_+_51795667 | 1.31 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr16_+_23975930 | 1.31 |
ENSDART00000147858
ENSDART00000144347 ENSDART00000115270 |
apoc4
|
apolipoprotein C-IV |
| chr18_+_28106139 | 1.31 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr14_+_23717165 | 1.29 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr23_-_36446307 | 1.27 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr15_+_21262917 | 1.26 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr14_+_25817628 | 1.26 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr5_-_51619742 | 1.24 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr20_+_29209926 | 1.23 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr3_-_56541723 | 1.22 |
ENSDART00000156398
ENSDART00000050576 ENSDART00000184874 |
si:ch211-189a21.1
cyth1a
|
si:ch211-189a21.1 cytohesin 1a |
| chr7_-_26532089 | 1.21 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr3_-_46811611 | 1.21 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr21_+_32820175 | 1.19 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr25_-_29087925 | 1.17 |
ENSDART00000171758
|
rpp25a
|
ribonuclease P and MRP subunit p25, a |
| chr19_+_22062202 | 1.17 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr12_-_35393211 | 1.15 |
ENSDART00000137139
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr9_+_25776971 | 1.15 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr10_-_34916208 | 1.14 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr9_+_19529951 | 1.13 |
ENSDART00000125416
|
pknox1.1
|
pbx/knotted 1 homeobox 1.1 |
| chr6_-_12275836 | 1.13 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr24_+_19591893 | 1.13 |
ENSDART00000152026
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr11_-_39044595 | 1.11 |
ENSDART00000065461
|
cldn19
|
claudin 19 |
| chr7_-_26467404 | 1.08 |
ENSDART00000170859
|
mblac1
|
metallo-beta-lactamase domain containing 1 |
| chr22_+_5176693 | 1.08 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr6_-_40713183 | 1.08 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr5_-_58112032 | 1.06 |
ENSDART00000016418
|
drd2b
|
dopamine receptor D2b |
| chr23_-_2901167 | 1.05 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr11_+_5588122 | 1.04 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr10_+_33393829 | 1.04 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr4_-_9891874 | 1.01 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr6_-_40922971 | 0.99 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr10_+_36026576 | 0.99 |
ENSDART00000193786
|
hmgb1a
|
high mobility group box 1a |
| chr17_+_10593398 | 0.98 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr17_+_3379673 | 0.97 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr14_-_7207961 | 0.96 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr14_-_33105434 | 0.96 |
ENSDART00000163795
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr17_+_30545895 | 0.95 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr15_-_22074315 | 0.95 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr17_-_8570257 | 0.94 |
ENSDART00000154713
ENSDART00000121488 |
fzd3b
|
frizzled class receptor 3b |
| chr7_+_36898850 | 0.93 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr22_-_8725768 | 0.92 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr24_-_26399623 | 0.92 |
ENSDART00000112317
|
zgc:194621
|
zgc:194621 |
| chr7_-_31759602 | 0.91 |
ENSDART00000113467
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr6_-_51386656 | 0.90 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr5_-_69004007 | 0.89 |
ENSDART00000137443
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr16_+_23976227 | 0.88 |
ENSDART00000193013
|
apoc4
|
apolipoprotein C-IV |
| chr1_+_10318089 | 0.88 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr12_-_14143344 | 0.88 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr9_-_43538328 | 0.87 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr12_-_29233738 | 0.87 |
ENSDART00000153175
|
si:ch211-214e3.5
|
si:ch211-214e3.5 |
| chr17_+_11675362 | 0.86 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.8 | 7.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 1.8 | 16.0 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 1.8 | 5.3 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.1 | 3.4 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.1 | 4.3 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 1.0 | 3.0 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.9 | 2.8 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.7 | 2.9 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.6 | 3.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.6 | 1.8 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.6 | 3.0 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.6 | 2.9 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.6 | 12.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.6 | 2.2 | GO:0010543 | regulation of platelet activation(GO:0010543) positive regulation of cell adhesion mediated by integrin(GO:0033630) regulation of platelet aggregation(GO:0090330) |
| 0.5 | 3.8 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.5 | 4.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.4 | 1.3 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.4 | 1.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.4 | 4.4 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.4 | 3.7 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.4 | 1.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 1.4 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.3 | 2.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.3 | 2.6 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.3 | 5.6 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.2 | 4.1 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.2 | 0.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 1.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.2 | 1.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.2 | 0.7 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.2 | 3.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 1.7 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.2 | 4.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 1.9 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 1.2 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.2 | 3.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 1.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.2 | 3.5 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.2 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 5.7 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 1.5 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.2 | 1.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.2 | 3.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.7 | GO:0019682 | glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 0.6 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 1.5 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 5.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.7 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.4 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 2.8 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 2.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.4 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 5.1 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.1 | 2.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 1.0 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 4.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 2.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 5.1 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.1 | 2.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 2.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.6 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 1.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.5 | GO:0060465 | pharynx development(GO:0060465) |
| 0.1 | 1.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.6 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 3.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 0.5 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 2.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 3.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.0 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 1.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 3.0 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 2.0 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.5 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 0.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 4.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.5 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.5 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.0 | 1.3 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.0 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.2 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 2.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 3.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.6 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 1.8 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.1 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 2.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.5 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.4 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.4 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 9.2 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 5.5 | GO:0099536 | synaptic signaling(GO:0099536) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 4.5 | GO:0000003 | reproduction(GO:0000003) |
| 0.0 | 3.1 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.4 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.5 | 3.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 3.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 1.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.4 | 3.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.4 | 1.5 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.4 | 3.7 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.3 | 1.8 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.3 | 2.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 2.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 3.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 7.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 4.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.4 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 6.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.0 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 1.0 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 2.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 3.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 4.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.4 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 6.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 7.7 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 0.3 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 1.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.4 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 2.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 5.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.5 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 2.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 7.5 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 6.7 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 5.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 12.9 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 3.4 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 4.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 3.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 2.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 4.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 6.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 1.8 | 7.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 1.8 | 5.3 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 1.5 | 22.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 1.4 | 4.1 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.9 | 2.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.6 | 3.0 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.5 | 3.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 2.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 1.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.4 | 1.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 1.7 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 7.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 6.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 2.0 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.3 | 3.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 2.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 3.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 4.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 0.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 2.5 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 1.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.0 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 3.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 1.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 1.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.7 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 3.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 8.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 1.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 5.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 2.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.3 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.5 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 3.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 4.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.9 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 5.9 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 2.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 3.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.7 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 3.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 2.1 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 4.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 5.5 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 3.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 2.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.6 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 1.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 3.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 3.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 10.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 2.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 9.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.9 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 6.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.6 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 2.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 3.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 1.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 5.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |