Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
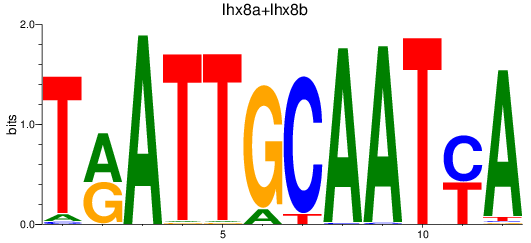
Results for lhx8a+lhx8b
Z-value: 0.83
Transcription factors associated with lhx8a+lhx8b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx8a
|
ENSDARG00000002330 | LIM homeobox 8a |
|
lhx8b
|
ENSDARG00000042145 | LIM homeobox 8b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx8a | dr11_v1_chr2_+_11205795_11205795 | 0.50 | 2.6e-07 | Click! |
| lhx8b | dr11_v1_chr8_-_17926814_17926814 | -0.10 | 3.4e-01 | Click! |
Activity profile of lhx8a+lhx8b motif
Sorted Z-values of lhx8a+lhx8b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_7546259 | 10.85 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr3_+_49021079 | 9.26 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr5_-_46505691 | 8.98 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr15_-_23645810 | 8.36 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr4_-_17785120 | 7.65 |
ENSDART00000024775
|
mybpc1
|
myosin binding protein C, slow type |
| chr11_+_25276748 | 7.33 |
ENSDART00000126211
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr17_-_22552678 | 6.67 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr11_+_6650966 | 5.85 |
ENSDART00000131236
|
si:dkey-246j7.1
|
si:dkey-246j7.1 |
| chr16_-_12319822 | 5.68 |
ENSDART00000127453
ENSDART00000184526 |
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr10_-_43568239 | 5.35 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr15_-_34567370 | 5.27 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr8_-_23612462 | 4.93 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr20_-_22476255 | 4.93 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr7_-_31441420 | 4.88 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr16_+_20926673 | 4.70 |
ENSDART00000009827
|
hoxa2b
|
homeobox A2b |
| chr5_-_23749348 | 4.67 |
ENSDART00000140766
|
gbgt1l2
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 2 |
| chr24_+_38306010 | 4.56 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr22_-_29586608 | 4.23 |
ENSDART00000059869
|
adra2a
|
adrenoceptor alpha 2A |
| chr6_+_40661703 | 4.04 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr18_+_8340886 | 3.88 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr5_+_51026563 | 3.88 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr13_-_33321058 | 3.86 |
ENSDART00000112084
|
si:dkey-71p21.13
|
si:dkey-71p21.13 |
| chr16_+_25535993 | 3.77 |
ENSDART00000077436
|
mylipb
|
myosin regulatory light chain interacting protein b |
| chr1_-_59077650 | 3.62 |
ENSDART00000043516
|
MFAP4 (1 of many)
|
si:zfos-2330d3.1 |
| chr1_-_58064738 | 3.62 |
ENSDART00000073778
|
caspb
|
caspase b |
| chr19_-_31042570 | 3.57 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr7_+_2228276 | 3.53 |
ENSDART00000064294
|
si:dkey-187j14.4
|
si:dkey-187j14.4 |
| chr15_+_32268790 | 3.53 |
ENSDART00000154457
|
fhdc4
|
FH2 domain containing 4 |
| chr22_+_6254194 | 3.47 |
ENSDART00000112388
ENSDART00000135176 |
rnasel4
|
ribonuclease like 4 |
| chr9_-_23383131 | 3.38 |
ENSDART00000101685
|
sh3bp4
|
SH3-domain binding protein 4 |
| chr25_+_31405266 | 3.35 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr5_-_65000312 | 3.33 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr22_-_23000815 | 3.25 |
ENSDART00000137111
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr22_-_17458070 | 3.23 |
ENSDART00000139658
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr6_-_32093830 | 3.19 |
ENSDART00000017695
|
foxd3
|
forkhead box D3 |
| chr3_-_32169754 | 3.14 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr18_-_7539166 | 3.09 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr21_-_25601648 | 3.08 |
ENSDART00000042578
|
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr21_+_15346363 | 3.07 |
ENSDART00000134167
|
si:dkey-11o15.7
|
si:dkey-11o15.7 |
| chr11_-_22599584 | 3.02 |
ENSDART00000014062
|
myog
|
myogenin |
| chr3_-_32603191 | 3.01 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr9_+_33417969 | 2.98 |
ENSDART00000024795
|
gpr34b
|
G protein-coupled receptor 34b |
| chr5_+_36654817 | 2.93 |
ENSDART00000131339
|
capns1a
|
calpain, small subunit 1 a |
| chr12_-_44196222 | 2.91 |
ENSDART00000171730
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr18_-_48558420 | 2.90 |
ENSDART00000058987
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr5_+_27897504 | 2.83 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr18_-_8877077 | 2.82 |
ENSDART00000137266
|
si:dkey-95h12.2
|
si:dkey-95h12.2 |
| chr22_-_15602593 | 2.81 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr8_+_13389115 | 2.75 |
ENSDART00000184428
ENSDART00000154266 ENSDART00000049469 |
jak3
|
Janus kinase 3 (a protein tyrosine kinase, leukocyte) |
| chr20_-_39273987 | 2.66 |
ENSDART00000127173
|
clu
|
clusterin |
| chr11_-_4175773 | 2.64 |
ENSDART00000042221
|
ccdc3b
|
coiled-coil domain containing 3b |
| chr7_-_22941472 | 2.62 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr5_+_32247310 | 2.47 |
ENSDART00000182649
|
myha
|
myosin, heavy chain a |
| chr20_-_39273505 | 2.46 |
ENSDART00000153114
|
clu
|
clusterin |
| chr5_+_51909740 | 2.42 |
ENSDART00000162541
|
thbs4a
|
thrombospondin 4a |
| chr7_+_53152108 | 2.41 |
ENSDART00000171350
|
cdh29
|
cadherin 29 |
| chr12_+_42574148 | 2.39 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr16_+_29514473 | 2.38 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr20_-_25436389 | 2.38 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr3_+_612432 | 2.37 |
ENSDART00000190912
|
CR792453.3
|
|
| chr20_+_16170848 | 2.32 |
ENSDART00000182115
|
zyg11
|
zyg-11 homolog (C. elegans) |
| chr15_+_33991928 | 2.26 |
ENSDART00000170177
|
vwde
|
von Willebrand factor D and EGF domains |
| chr22_-_15602760 | 2.25 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr3_+_464127 | 2.23 |
ENSDART00000169510
ENSDART00000144886 |
dicp1.1
|
diverse immunoglobulin domain-containing protein 1.1 |
| chr3_-_37773800 | 2.22 |
ENSDART00000151341
|
baxa
|
BCL2 associated X, apoptosis regulator a |
| chr20_+_6535438 | 2.22 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr11_+_21050326 | 2.19 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr1_+_52929185 | 2.18 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr2_-_32384683 | 2.17 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr4_-_25812329 | 2.17 |
ENSDART00000146658
|
tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr16_+_45739193 | 2.15 |
ENSDART00000184852
ENSDART00000156851 ENSDART00000154704 |
paqr6
|
progestin and adipoQ receptor family member VI |
| chr18_-_7539469 | 2.15 |
ENSDART00000101296
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr18_+_924949 | 2.11 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr6_+_19948043 | 2.11 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr18_-_2707218 | 2.00 |
ENSDART00000175226
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr1_-_40735381 | 1.98 |
ENSDART00000093269
|
zgc:153642
|
zgc:153642 |
| chr13_+_32148338 | 1.98 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr20_+_25225112 | 1.87 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr5_+_33339762 | 1.84 |
ENSDART00000026085
|
ptges
|
prostaglandin E synthase |
| chr12_+_18782821 | 1.82 |
ENSDART00000152918
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr25_+_7591293 | 1.81 |
ENSDART00000130416
|
FO704779.1
|
|
| chr1_+_1904419 | 1.79 |
ENSDART00000142874
|
si:ch211-132g1.4
|
si:ch211-132g1.4 |
| chr24_+_19578935 | 1.78 |
ENSDART00000137175
|
sulf1
|
sulfatase 1 |
| chr11_-_43200994 | 1.76 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr4_+_9536860 | 1.75 |
ENSDART00000130083
|
lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr10_-_44482911 | 1.74 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr13_-_9598320 | 1.74 |
ENSDART00000184613
|
cpxm1a
|
carboxypeptidase X (M14 family), member 1a |
| chr13_-_8892514 | 1.67 |
ENSDART00000144553
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr13_-_1085961 | 1.62 |
ENSDART00000171872
ENSDART00000166598 |
plek
|
pleckstrin |
| chr20_-_49681850 | 1.59 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr19_-_30811161 | 1.58 |
ENSDART00000103524
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr16_-_35329803 | 1.57 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr3_+_34120191 | 1.54 |
ENSDART00000020017
ENSDART00000151700 |
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr7_+_20512419 | 1.47 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr12_+_9551667 | 1.45 |
ENSDART00000048493
ENSDART00000166178 |
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr4_+_37800207 | 1.43 |
ENSDART00000189015
|
si:ch211-271g18.1
|
si:ch211-271g18.1 |
| chr7_-_5316901 | 1.42 |
ENSDART00000181505
ENSDART00000124367 |
si:cabz01074946.1
|
si:cabz01074946.1 |
| chr20_-_7080427 | 1.41 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr7_+_41887429 | 1.40 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr17_-_30652738 | 1.38 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr1_+_58221372 | 1.31 |
ENSDART00000138467
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr20_+_34151670 | 1.30 |
ENSDART00000152870
ENSDART00000010329 ENSDART00000145852 |
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr9_+_42607138 | 1.30 |
ENSDART00000138133
ENSDART00000002027 |
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr8_+_23726244 | 1.29 |
ENSDART00000132734
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr25_-_348784 | 1.28 |
ENSDART00000059514
|
prickle1a
|
prickle homolog 1a |
| chr1_-_55873178 | 1.27 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr20_-_38446891 | 1.26 |
ENSDART00000192013
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr4_+_70015788 | 1.24 |
ENSDART00000161133
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr13_-_15994419 | 1.21 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr8_-_2289264 | 1.18 |
ENSDART00000189397
|
plat
|
plasminogen activator, tissue |
| chr1_-_8140763 | 1.15 |
ENSDART00000091508
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr9_+_13230214 | 1.13 |
ENSDART00000017932
ENSDART00000179850 |
carf
|
calcium responsive transcription factor |
| chr19_-_40191358 | 1.11 |
ENSDART00000183919
|
grn1
|
granulin 1 |
| chr21_+_5531138 | 1.08 |
ENSDART00000163825
|
ly6m6
|
lymphocyte antigen 6 family member M6 |
| chr7_+_69653981 | 1.06 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr4_+_612363 | 1.05 |
ENSDART00000049154
|
pthlha
|
parathyroid hormone-like hormone a |
| chr9_+_42606681 | 1.00 |
ENSDART00000191186
|
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr20_+_18551657 | 0.99 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr16_-_9802449 | 0.99 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr18_+_31410652 | 0.99 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr22_+_38164486 | 0.98 |
ENSDART00000137521
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr4_+_42878556 | 0.94 |
ENSDART00000171047
|
si:zfos-451d2.2
|
si:zfos-451d2.2 |
| chr19_+_816208 | 0.94 |
ENSDART00000093304
|
nrm
|
nurim |
| chr13_-_33667922 | 0.93 |
ENSDART00000141631
|
si:dkey-76k16.5
|
si:dkey-76k16.5 |
| chr25_+_4635355 | 0.92 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr5_+_1624359 | 0.91 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr9_-_21918963 | 0.91 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr14_-_23801389 | 0.91 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr1_-_58036509 | 0.91 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr9_+_25776971 | 0.90 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr9_+_9858935 | 0.89 |
ENSDART00000137206
|
si:ch1073-394i4.1
|
si:ch1073-394i4.1 |
| chr18_+_6319447 | 0.88 |
ENSDART00000143740
|
ptprq
|
protein tyrosine phosphatase, receptor type, Q |
| chr8_+_10823069 | 0.86 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr4_-_4751981 | 0.85 |
ENSDART00000147436
ENSDART00000092984 ENSDART00000158466 |
creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr9_-_16853462 | 0.85 |
ENSDART00000160273
|
CT573248.2
|
|
| chr16_-_36979592 | 0.84 |
ENSDART00000168443
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr9_+_25775816 | 0.84 |
ENSDART00000127834
ENSDART00000189994 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr24_+_20920563 | 0.83 |
ENSDART00000037224
|
cst14a.2
|
cystatin 14a, tandem duplicate 2 |
| chr17_+_24800332 | 0.83 |
ENSDART00000156504
|
si:dkey-74h17.3
|
si:dkey-74h17.3 |
| chr19_+_10339538 | 0.82 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr7_-_64770456 | 0.82 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr5_+_27525477 | 0.80 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr15_-_24960730 | 0.80 |
ENSDART00000109990
ENSDART00000186706 |
abhd15a
|
abhydrolase domain containing 15a |
| chr3_-_45281350 | 0.79 |
ENSDART00000020168
|
kctd5a
|
potassium channel tetramerization domain containing 5a |
| chr4_+_65614183 | 0.78 |
ENSDART00000180387
|
si:dkey-205i10.1
|
si:dkey-205i10.1 |
| chr18_-_21105968 | 0.74 |
ENSDART00000100791
|
si:dkey-12e7.1
|
si:dkey-12e7.1 |
| chr24_-_32025637 | 0.73 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr16_-_35427060 | 0.72 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr23_-_43486714 | 0.71 |
ENSDART00000169726
|
e2f1
|
E2F transcription factor 1 |
| chr18_+_30370559 | 0.68 |
ENSDART00000165450
|
gse1
|
Gse1 coiled-coil protein |
| chr15_-_41503392 | 0.68 |
ENSDART00000169351
|
si:dkey-230i18.2
|
si:dkey-230i18.2 |
| chr9_+_6079528 | 0.64 |
ENSDART00000142167
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr24_+_12945803 | 0.64 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr1_+_30100257 | 0.64 |
ENSDART00000134311
|
vps8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr11_+_44236183 | 0.64 |
ENSDART00000193470
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr6_-_12456077 | 0.63 |
ENSDART00000190107
|
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr22_+_9069081 | 0.63 |
ENSDART00000187842
|
BX248395.1
|
|
| chr11_-_891674 | 0.62 |
ENSDART00000172904
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr18_+_26899316 | 0.61 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr23_+_26079467 | 0.61 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr5_+_13326765 | 0.59 |
ENSDART00000090851
|
ydjc
|
YdjC chitooligosaccharide deacetylase homolog |
| chr23_+_29358188 | 0.59 |
ENSDART00000189242
|
tardbpl
|
TAR DNA binding protein, like |
| chr3_-_52674089 | 0.59 |
ENSDART00000154260
ENSDART00000125455 |
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr15_+_36966369 | 0.59 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr12_-_4370585 | 0.58 |
ENSDART00000129502
|
CA4 (1 of many)
|
si:ch211-173d10.4 |
| chr1_-_23268569 | 0.58 |
ENSDART00000143948
|
rfc1
|
replication factor C (activator 1) 1 |
| chr9_-_2892250 | 0.56 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr17_-_33412868 | 0.55 |
ENSDART00000187521
|
BX323819.1
|
|
| chr18_-_26811959 | 0.55 |
ENSDART00000086250
|
znf592
|
zinc finger protein 592 |
| chr13_+_23162447 | 0.54 |
ENSDART00000180209
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr4_-_71469799 | 0.54 |
ENSDART00000187156
|
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr10_+_20364009 | 0.53 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr19_-_11950434 | 0.53 |
ENSDART00000024861
|
cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr10_+_24631976 | 0.51 |
ENSDART00000181928
|
slc46a3
|
solute carrier family 46, member 3 |
| chr5_-_15494164 | 0.50 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr23_-_29668286 | 0.49 |
ENSDART00000129248
|
clstn1
|
calsyntenin 1 |
| chr16_-_39195318 | 0.48 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr7_-_30280934 | 0.48 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr4_+_14727018 | 0.46 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr4_+_14727212 | 0.46 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr16_+_49088698 | 0.45 |
ENSDART00000182037
|
plcl2
|
phospholipase C like 2 |
| chr16_-_25535430 | 0.45 |
ENSDART00000077420
|
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr20_+_492529 | 0.44 |
ENSDART00000141709
|
FO904980.1
|
|
| chr21_-_34926619 | 0.44 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr4_+_15605844 | 0.43 |
ENSDART00000101619
ENSDART00000021384 |
exoc4
|
exocyst complex component 4 |
| chr24_+_119680 | 0.42 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr4_-_25515154 | 0.42 |
ENSDART00000186524
|
rbm17
|
RNA binding motif protein 17 |
| chr17_-_2386569 | 0.42 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr20_-_51727860 | 0.41 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr19_+_366034 | 0.40 |
ENSDART00000093383
|
vps72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr8_+_26254903 | 0.39 |
ENSDART00000113763
|
slc26a6
|
solute carrier family 26, member 6 |
| chr22_+_1313046 | 0.39 |
ENSDART00000170421
|
si:ch73-138e16.6
|
si:ch73-138e16.6 |
| chr8_-_19216180 | 0.38 |
ENSDART00000146162
|
si:ch73-222f22.2
|
si:ch73-222f22.2 |
| chr9_-_16877456 | 0.38 |
ENSDART00000161105
ENSDART00000160869 |
fbxl3a
|
F-box and leucine-rich repeat protein 3a |
| chr4_-_22311610 | 0.38 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr15_-_34865952 | 0.38 |
ENSDART00000186868
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr17_-_8173380 | 0.37 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr21_+_27302752 | 0.37 |
ENSDART00000012855
|
sart1
|
SART1, U4/U6.U5 tri-snRNP-associated protein 1 |
| chr20_-_894374 | 0.37 |
ENSDART00000104725
ENSDART00000142361 |
snx14
|
sorting nexin 14 |
| chr1_-_21538673 | 0.37 |
ENSDART00000131257
|
wdr19
|
WD repeat domain 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx8a+lhx8b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 1.4 | 5.7 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 1.2 | 4.9 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 1.0 | 9.9 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 1.0 | 4.9 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.9 | 2.8 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.8 | 4.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.7 | 2.0 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.6 | 8.4 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.6 | 3.2 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.6 | 1.9 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.6 | 3.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.6 | 2.8 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.5 | 4.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.4 | 3.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 1.1 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.4 | 1.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.3 | 1.2 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.3 | 0.9 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.3 | 2.2 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.3 | 4.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 0.9 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.2 | 1.8 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 3.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.2 | 6.1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.2 | 0.7 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 5.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 3.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 0.6 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 3.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 3.3 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 0.9 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.4 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.1 | 2.2 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 1.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 1.5 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 5.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 7.7 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 0.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.0 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.3 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 0.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 9.0 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 5.1 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.1 | 1.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 1.2 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 1.6 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 3.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 3.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.4 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 0.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 1.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 4.2 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 1.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.6 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.5 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 1.7 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 4.6 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 6.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 2.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.6 | GO:0010950 | positive regulation of endopeptidase activity(GO:0010950) positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 1.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 2.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 2.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.9 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 2.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.6 | GO:0039022 | pronephric duct development(GO:0039022) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0002312 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.0 | 0.3 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.4 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.5 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 1.9 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 1.9 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.3 | GO:0043489 | RNA stabilization(GO:0043489) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.3 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 3.4 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.3 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0043034 | costamere(GO:0043034) |
| 0.9 | 3.6 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.5 | 5.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.5 | 4.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 2.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.4 | 1.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.7 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 1.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 2.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 6.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.6 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 4.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 5.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 5.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.8 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 2.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 4.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 8.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0031968 | outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 12.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.7 | 4.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.7 | 5.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.6 | 8.4 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.6 | 1.9 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.6 | 2.8 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.6 | 3.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 4.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 7.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 4.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 1.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 2.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 2.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 0.9 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 3.3 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 3.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.3 | 3.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 1.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 9.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 5.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 0.7 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 2.7 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 1.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 1.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.6 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.8 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.1 | 1.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 5.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.9 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 0.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.3 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 2.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.0 | 1.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 3.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.9 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.3 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 7.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.5 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 2.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 4.7 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 8.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 3.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 15.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.2 | 1.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 8.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 4.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 2.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 5.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 6.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.4 | 10.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 4.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 3.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 2.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 4.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 6.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 4.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |