Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
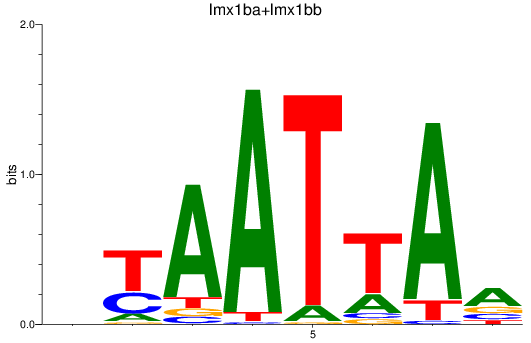
Results for lmx1ba+lmx1bb_lmx1al
Z-value: 0.57
Transcription factors associated with lmx1ba+lmx1bb_lmx1al
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lmx1bb
|
ENSDARG00000068365 | LIM homeobox transcription factor 1, beta b |
|
lmx1ba
|
ENSDARG00000104815 | LIM homeobox transcription factor 1, beta a |
|
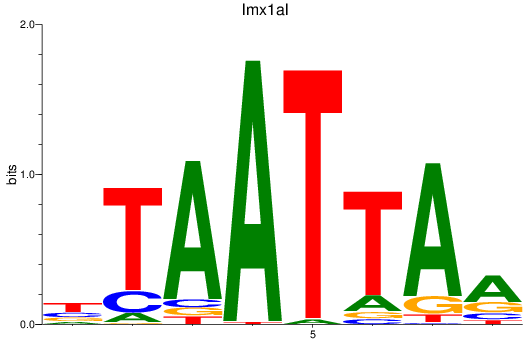
lmx1al
|
ENSDARG00000077915 | LIM homeobox transcription factor 1, alpha-like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lmx1al | dr11_v1_chr3_+_52806347_52806441 | -0.27 | 9.2e-03 | Click! |
| lmx1bb | dr11_v1_chr8_-_33381913_33381913 | -0.20 | 5.7e-02 | Click! |
| lmx1ba | dr11_v1_chr5_-_5035683_5035683 | 0.11 | 3.0e-01 | Click! |
Activity profile of lmx1ba+lmx1bb_lmx1al motif
Sorted Z-values of lmx1ba+lmx1bb_lmx1al motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23431189 | 7.39 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr16_+_23961276 | 6.92 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr12_-_35830625 | 6.85 |
ENSDART00000180028
|
CU459056.1
|
|
| chr20_-_40755614 | 6.22 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr22_+_19552987 | 6.11 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr16_+_29509133 | 5.41 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr25_-_3867990 | 5.19 |
ENSDART00000075663
|
cracr2b
|
calcium release activated channel regulator 2B |
| chr17_+_28103675 | 5.02 |
ENSDART00000188078
|
zgc:91908
|
zgc:91908 |
| chr16_+_26777473 | 4.65 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr7_+_25033924 | 4.31 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr10_+_21867307 | 3.89 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr6_-_43283122 | 3.69 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr24_-_38110779 | 3.68 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr2_+_33326522 | 3.59 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr3_-_15081874 | 3.57 |
ENSDART00000192532
|
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr14_-_33481428 | 3.53 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr18_+_15644559 | 3.48 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr2_+_20793982 | 3.47 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr8_+_11425048 | 3.41 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr7_+_20503344 | 3.39 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr13_-_31017960 | 3.36 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr20_-_22476255 | 3.26 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr8_+_6576940 | 3.14 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr24_-_35561672 | 3.11 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr11_-_45138857 | 2.92 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr15_+_36457888 | 2.90 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr6_+_41191482 | 2.90 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr19_-_13808630 | 2.85 |
ENSDART00000166895
ENSDART00000187670 |
ctgfb
|
connective tissue growth factor b |
| chr24_-_2450597 | 2.81 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr1_-_10071422 | 2.70 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr16_+_33902006 | 2.68 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr1_+_44173245 | 2.67 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr17_-_24575893 | 2.65 |
ENSDART00000141914
|
aftphb
|
aftiphilin b |
| chr13_+_27232848 | 2.63 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr15_-_21877726 | 2.63 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr19_+_43297546 | 2.54 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr13_+_25720969 | 2.48 |
ENSDART00000046050
|
pcbd1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr1_+_45056371 | 2.39 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr6_+_40922572 | 2.33 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr3_+_27798094 | 2.32 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr24_+_19415124 | 2.28 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr22_-_15578402 | 2.26 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr15_-_43284021 | 2.26 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr19_-_5699703 | 2.25 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr22_-_10121880 | 2.18 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr18_-_19456269 | 2.18 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr23_-_31913069 | 2.15 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr1_-_10821584 | 2.14 |
ENSDART00000167452
ENSDART00000162137 |
crfb15
|
cytokine receptor family member B15 |
| chr6_-_18228358 | 2.12 |
ENSDART00000167937
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr22_+_1170294 | 2.11 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr18_+_3037998 | 2.10 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr21_+_25777425 | 2.06 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr13_+_28747934 | 2.05 |
ENSDART00000160176
ENSDART00000101633 ENSDART00000136318 |
prom2
|
prominin 2 |
| chr23_+_39695827 | 2.03 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr17_+_16090436 | 2.03 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr14_+_33329420 | 2.01 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr22_-_17653143 | 2.00 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr17_-_41798856 | 1.98 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr8_-_20230559 | 1.98 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr20_-_23426339 | 1.97 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr7_-_26306546 | 1.97 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr16_+_23913943 | 1.96 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr18_+_7073130 | 1.89 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr17_-_21884323 | 1.88 |
ENSDART00000155574
|
cuzd1.1
|
CUB and zona pellucida-like domains 1, tandem duplicate 1 |
| chr3_-_16719244 | 1.85 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr25_+_18475032 | 1.83 |
ENSDART00000073564
|
tes
|
testis derived transcript (3 LIM domains) |
| chr4_-_9891874 | 1.80 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr8_-_12867128 | 1.80 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr16_-_29387215 | 1.79 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr10_-_21362071 | 1.70 |
ENSDART00000125167
|
avd
|
avidin |
| chr8_+_16758304 | 1.70 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr11_+_37250839 | 1.69 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr23_-_31913231 | 1.69 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr3_-_61162750 | 1.66 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr1_+_9966384 | 1.64 |
ENSDART00000132607
|
si:dkeyp-75b4.8
|
si:dkeyp-75b4.8 |
| chr5_+_36850650 | 1.63 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr13_-_31008275 | 1.59 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr10_+_11261576 | 1.59 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr6_+_3343834 | 1.56 |
ENSDART00000160290
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr9_+_8396755 | 1.52 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr10_+_29850330 | 1.49 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr24_+_13316737 | 1.48 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr14_+_23717165 | 1.48 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr25_-_21031007 | 1.47 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr16_-_17347727 | 1.47 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr2_-_26596794 | 1.45 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr10_-_15340362 | 1.45 |
ENSDART00000148119
ENSDART00000127277 ENSDART00000154037 ENSDART00000189109 |
pum3
|
pumilio RNA-binding family member 3 |
| chr19_+_43119014 | 1.44 |
ENSDART00000023156
|
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr9_-_9415000 | 1.42 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr10_-_21362320 | 1.42 |
ENSDART00000189789
|
avd
|
avidin |
| chr20_+_46492175 | 1.42 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr1_-_16665044 | 1.41 |
ENSDART00000040434
|
asah1b
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1b |
| chr18_-_6982499 | 1.41 |
ENSDART00000101525
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr13_+_24750078 | 1.40 |
ENSDART00000021053
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr11_-_38533505 | 1.39 |
ENSDART00000113894
|
slc45a3
|
solute carrier family 45, member 3 |
| chr18_+_20560616 | 1.39 |
ENSDART00000136710
ENSDART00000151974 ENSDART00000121699 ENSDART00000040074 |
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr14_+_35428152 | 1.39 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr16_-_42056137 | 1.39 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr22_-_17652914 | 1.37 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr1_+_52392511 | 1.37 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr8_-_23612462 | 1.35 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr8_-_12867434 | 1.35 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr10_-_43771447 | 1.34 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr10_+_8885769 | 1.31 |
ENSDART00000139466
|
itga2.2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 2 |
| chr20_-_52902693 | 1.30 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr24_-_6078222 | 1.25 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr2_+_41526904 | 1.23 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr7_+_56577522 | 1.22 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr21_-_35419486 | 1.19 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr5_+_6954162 | 1.19 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr17_+_25481466 | 1.18 |
ENSDART00000139451
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr3_-_32337653 | 1.18 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr6_+_39098397 | 1.17 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr19_+_30884960 | 1.16 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr3_+_17537352 | 1.15 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr7_+_56577906 | 1.12 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr18_-_48550426 | 1.12 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr22_+_19220459 | 1.12 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr14_-_22113600 | 1.12 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr15_-_23376541 | 1.10 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr24_-_40860603 | 1.09 |
ENSDART00000188032
|
CU633479.7
|
|
| chr7_-_71389375 | 1.09 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr8_-_44378391 | 1.06 |
ENSDART00000158784
ENSDART00000098138 |
si:busm1-228j01.6
|
si:busm1-228j01.6 |
| chr6_+_45918981 | 1.06 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr8_+_23152244 | 1.06 |
ENSDART00000036801
ENSDART00000184298 |
slc17a9a
|
solute carrier family 17 (vesicular nucleotide transporter), member 9a |
| chr24_-_30862168 | 1.05 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr5_+_33519943 | 1.04 |
ENSDART00000131316
|
ms4a17c.1
|
membrane-spanning 4-domains, subfamily A, member 17C.1 |
| chr18_+_19456648 | 1.04 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr18_+_2228737 | 1.01 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr19_+_41464870 | 1.01 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr16_+_23087326 | 0.99 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr16_+_23303859 | 0.98 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr17_-_26867725 | 0.98 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr16_-_16761164 | 0.97 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr24_-_19719240 | 0.97 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr11_+_35171406 | 0.96 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr20_-_37813863 | 0.96 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr8_-_20230802 | 0.94 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr8_+_45334255 | 0.93 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr16_-_38609146 | 0.93 |
ENSDART00000144651
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr1_-_43905252 | 0.93 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr2_+_36112273 | 0.92 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr19_+_30884706 | 0.91 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr23_+_31913292 | 0.91 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr24_-_27452488 | 0.90 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr3_+_46635527 | 0.89 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr3_+_1015867 | 0.89 |
ENSDART00000109912
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr1_+_52929185 | 0.88 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr3_+_17933132 | 0.86 |
ENSDART00000104299
ENSDART00000162144 ENSDART00000162242 ENSDART00000166289 ENSDART00000171101 ENSDART00000164853 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr9_+_24008879 | 0.85 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr11_-_41853874 | 0.85 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr21_+_5580948 | 0.85 |
ENSDART00000160373
|
ly6m7
|
lymphocyte antigen 6 family member M7 |
| chr5_+_66433287 | 0.83 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr2_+_19522082 | 0.82 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr8_-_37249813 | 0.82 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr10_+_25726694 | 0.82 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr2_-_55298075 | 0.82 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr3_-_21094437 | 0.81 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr15_-_20400423 | 0.81 |
ENSDART00000081290
ENSDART00000156813 ENSDART00000192427 |
rnaset2l
|
ribonuclease T2, like |
| chr8_-_25034411 | 0.80 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr6_-_16717878 | 0.80 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr2_+_29842878 | 0.79 |
ENSDART00000131376
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr24_-_4450238 | 0.79 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr24_+_16985181 | 0.79 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr13_+_35339182 | 0.77 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr2_+_36721357 | 0.77 |
ENSDART00000019063
|
pdcd10b
|
programmed cell death 10b |
| chr20_-_43663494 | 0.76 |
ENSDART00000144564
|
BX470188.1
|
|
| chr6_-_20952187 | 0.76 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr1_-_18811517 | 0.76 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr23_-_24394719 | 0.75 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr25_-_18435481 | 0.75 |
ENSDART00000004771
|
poc1b
|
POC1 centriolar protein B |
| chr10_-_26512993 | 0.75 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr3_-_13921173 | 0.74 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr12_+_47698356 | 0.74 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr21_+_42717424 | 0.74 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr5_-_69620722 | 0.73 |
ENSDART00000097248
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr16_-_29528198 | 0.73 |
ENSDART00000150028
|
onecutl
|
one cut domain, family member, like |
| chr16_-_51299061 | 0.72 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr10_+_6884627 | 0.71 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr6_+_3280939 | 0.71 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr9_+_50001746 | 0.71 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr5_+_35786141 | 0.71 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr2_-_37140423 | 0.70 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr1_-_5455498 | 0.70 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr8_-_14126646 | 0.66 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr9_+_34641237 | 0.66 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr8_+_25034544 | 0.66 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr13_-_45155792 | 0.65 |
ENSDART00000163556
|
runx3
|
runt-related transcription factor 3 |
| chr2_-_38363017 | 0.65 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr20_-_6532462 | 0.65 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr10_-_25217347 | 0.64 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr9_+_37329036 | 0.64 |
ENSDART00000131756
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr10_+_7593185 | 0.64 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr3_+_17933553 | 0.64 |
ENSDART00000167731
ENSDART00000165644 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr1_-_26444075 | 0.64 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr11_+_18873619 | 0.63 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr5_-_7199998 | 0.62 |
ENSDART00000167316
|
CABZ01087502.1
|
|
| chr24_-_40744672 | 0.62 |
ENSDART00000160672
|
CU633479.1
|
|
| chr12_+_16168342 | 0.61 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr5_+_25733774 | 0.58 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
Network of associatons between targets according to the STRING database.
First level regulatory network of lmx1ba+lmx1bb_lmx1al
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 1.2 | 3.6 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.7 | 3.3 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.6 | 2.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.5 | 3.8 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.5 | 2.6 | GO:0034380 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) high-density lipoprotein particle assembly(GO:0034380) |
| 0.5 | 1.4 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.5 | 1.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.5 | 1.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.4 | 1.3 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.4 | 2.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 2.4 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.3 | 1.4 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.3 | 3.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.3 | 1.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.3 | 1.2 | GO:0042755 | eating behavior(GO:0042755) |
| 0.3 | 2.3 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.3 | 0.8 | GO:0060898 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.3 | 0.8 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.2 | 2.5 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 1.5 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.2 | 3.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 3.5 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 0.8 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 0.8 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 0.6 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.2 | 0.8 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 3.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.4 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 2.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 2.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.4 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.7 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 1.0 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 2.1 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.6 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 1.1 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.3 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.1 | 1.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.7 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.8 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 1.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.6 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 1.0 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 0.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 1.0 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.1 | 0.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.3 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 2.3 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 0.9 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.4 | GO:0070265 | necrotic cell death(GO:0070265) programmed necrotic cell death(GO:0097300) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 2.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.5 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.7 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 4.3 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 2.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 3.4 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 3.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.2 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 2.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 1.0 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 2.4 | GO:0036293 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) |
| 0.0 | 0.7 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.5 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 2.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 2.6 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 4.3 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.0 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 0.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 1.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 1.5 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 7.2 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 2.6 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.5 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.1 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.7 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.4 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 1.5 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.0 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) negative regulation of nuclear division(GO:0051784) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.0 | GO:0009080 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.2 | 3.5 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.9 | 2.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 2.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 1.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 1.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.3 | 1.0 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.3 | 1.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 0.8 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.3 | 2.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.3 | 0.8 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.2 | 1.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 2.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 3.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.9 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.2 | 0.5 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.2 | 0.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.4 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 9.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 3.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 2.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 12.8 | GO:0000323 | lytic vacuole(GO:0000323) |
| 0.0 | 2.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.4 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.1 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 1.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 3.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 5.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0005795 | Golgi stack(GO:0005795) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.8 | 3.3 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.6 | 3.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.6 | 3.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.5 | 2.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.5 | 1.5 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.4 | 3.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 1.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.4 | 1.4 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.3 | 9.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 1.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 0.8 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.2 | 1.6 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 2.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.6 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 6.1 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.2 | 1.4 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 1.9 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.2 | 2.1 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 3.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 2.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 3.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 3.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 3.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 5.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 2.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.6 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.0 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.1 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 5.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 4.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 5.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.0 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 3.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 2.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 3.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 6.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 2.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.3 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 3.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 2.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 3.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 2.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 3.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 3.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 3.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 3.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 3.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 2.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 1.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 3.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 2.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 3.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |