Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
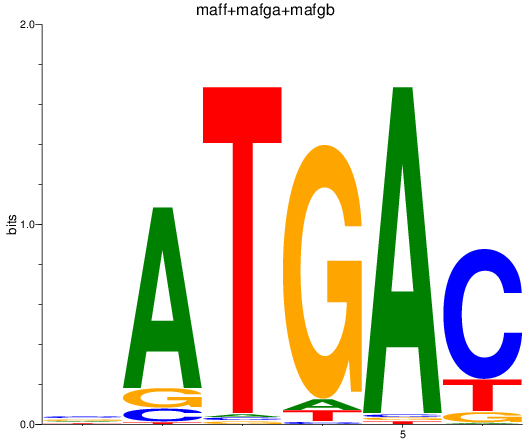
Results for maff+mafga+mafgb
Z-value: 0.90
Transcription factors associated with maff+mafga+mafgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafga
|
ENSDARG00000018109 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ga |
|
maff
|
ENSDARG00000028957 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
|
mafgb
|
ENSDARG00000100097 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
|
maff
|
ENSDARG00000111050 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafga | dr11_v1_chr3_-_55404985_55404985 | 0.50 | 2.5e-07 | Click! |
| mafgb | dr11_v1_chr11_+_45300669_45300669 | 0.45 | 4.5e-06 | Click! |
| maff | dr11_v1_chr12_-_19346678_19346678 | -0.13 | 2.0e-01 | Click! |
Activity profile of maff+mafga+mafgb motif
Sorted Z-values of maff+mafga+mafgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_32526799 | 16.88 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr2_-_36040820 | 7.67 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr6_-_35446110 | 7.63 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr19_-_19339285 | 7.44 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr6_+_27667359 | 7.30 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr14_-_30490763 | 6.68 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr3_+_15907297 | 6.19 |
ENSDART00000139206
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr20_+_27393668 | 6.08 |
ENSDART00000005473
|
tmem179
|
transmembrane protein 179 |
| chr7_+_25059845 | 6.01 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr9_+_27411502 | 5.95 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr15_-_44512461 | 5.72 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr16_+_10777116 | 5.64 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr16_-_29452039 | 5.62 |
ENSDART00000148960
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr12_-_10300101 | 5.37 |
ENSDART00000126428
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr20_+_20637866 | 5.25 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr3_+_15907458 | 5.19 |
ENSDART00000163525
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr16_-_12173554 | 5.07 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr6_-_31348999 | 5.02 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr2_+_20332044 | 5.00 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr19_+_25649626 | 4.98 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr23_-_27345425 | 4.95 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr21_-_43606502 | 4.67 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr3_+_31933893 | 4.65 |
ENSDART00000146509
ENSDART00000139644 |
lin7b
|
lin-7 homolog B (C. elegans) |
| chr23_-_4915118 | 4.61 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr10_-_27049170 | 4.56 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr3_-_39171968 | 4.55 |
ENSDART00000154494
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr10_+_29698467 | 4.25 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr21_-_45613564 | 4.13 |
ENSDART00000160324
|
LO018363.1
|
|
| chr21_+_9666347 | 4.13 |
ENSDART00000163853
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr14_-_2355833 | 4.11 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr5_+_23118470 | 4.01 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr20_+_20638034 | 3.99 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr2_-_24996441 | 3.98 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr8_+_14792830 | 3.96 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr8_+_53452681 | 3.96 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr13_+_28675686 | 3.95 |
ENSDART00000027213
|
inaa
|
internexin neuronal intermediate filament protein, alpha a |
| chr7_+_67486807 | 3.93 |
ENSDART00000159989
|
cpne7
|
copine VII |
| chr6_+_34512313 | 3.89 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr7_-_24491614 | 3.87 |
ENSDART00000131063
|
si:dkeyp-75h12.5
|
si:dkeyp-75h12.5 |
| chr19_-_9867001 | 3.83 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr21_+_18353703 | 3.80 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr13_+_24280380 | 3.77 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr7_-_57933736 | 3.72 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr8_+_39607466 | 3.71 |
ENSDART00000097427
|
msi1
|
musashi RNA-binding protein 1 |
| chr17_+_27134806 | 3.67 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr5_+_65491390 | 3.67 |
ENSDART00000159921
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr21_+_10794914 | 3.63 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr7_-_18730474 | 3.58 |
ENSDART00000170374
|
si:dkey-38l22.2
|
si:dkey-38l22.2 |
| chr21_-_24632778 | 3.57 |
ENSDART00000132533
ENSDART00000058370 |
arhgap32b
|
Rho GTPase activating protein 32b |
| chr3_-_32320537 | 3.53 |
ENSDART00000113550
ENSDART00000168483 |
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr7_+_23515966 | 3.49 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr6_-_10780698 | 3.44 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr2_+_23222939 | 3.43 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr2_+_20331445 | 3.43 |
ENSDART00000186880
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr13_-_36545258 | 3.40 |
ENSDART00000186171
|
FP103009.1
|
|
| chr18_+_7286788 | 3.36 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr7_-_69983948 | 3.30 |
ENSDART00000185827
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr6_+_52064304 | 3.30 |
ENSDART00000153468
|
abraa
|
actin binding Rho activating protein a |
| chr13_+_51579851 | 3.29 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr24_-_32408404 | 3.27 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr23_+_216012 | 3.26 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr9_-_4506819 | 3.25 |
ENSDART00000113975
|
kcnj3a
|
potassium inwardly-rectifying channel, subfamily J, member 3a |
| chr24_+_32472155 | 3.23 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr17_+_12698532 | 3.20 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr23_-_29502287 | 3.18 |
ENSDART00000141075
ENSDART00000053807 |
kif1b
|
kinesin family member 1B |
| chr1_-_10048514 | 3.17 |
ENSDART00000125358
ENSDART00000054835 |
rnf175
|
ring finger protein 175 |
| chr17_-_22062364 | 3.17 |
ENSDART00000114470
|
ttbk1b
|
tau tubulin kinase 1b |
| chr20_-_51917771 | 3.15 |
ENSDART00000147240
|
dusp10
|
dual specificity phosphatase 10 |
| chr21_+_20901505 | 3.13 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr21_-_28340977 | 3.06 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr5_+_27137473 | 3.05 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr2_+_36862473 | 3.03 |
ENSDART00000135624
|
si:dkey-193b15.8
|
si:dkey-193b15.8 |
| chr10_-_27009413 | 2.90 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr22_+_3238474 | 2.88 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr21_+_30194904 | 2.86 |
ENSDART00000137023
ENSDART00000078403 |
BRD8
|
si:ch211-59d17.3 |
| chr19_-_13286722 | 2.84 |
ENSDART00000168296
ENSDART00000158330 |
zfpm2b
|
zinc finger protein, FOG family member 2b |
| chr14_+_25464681 | 2.84 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr11_-_18791834 | 2.82 |
ENSDART00000156431
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr9_-_18424844 | 2.77 |
ENSDART00000154351
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr9_-_42861080 | 2.76 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr5_-_32158198 | 2.72 |
ENSDART00000187215
ENSDART00000137503 |
m17
|
IL-6 subfamily cytokine M17 |
| chr11_+_30729745 | 2.72 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr3_-_60142530 | 2.64 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr5_-_21422390 | 2.63 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr20_+_46206998 | 2.59 |
ENSDART00000074482
|
taar15
|
trace amine associated receptor 15 |
| chr3_-_32169754 | 2.58 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr10_+_42733210 | 2.58 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr7_+_42206543 | 2.57 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr2_-_32356539 | 2.56 |
ENSDART00000169316
|
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr20_-_27864964 | 2.54 |
ENSDART00000153311
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr10_+_21758811 | 2.53 |
ENSDART00000188827
|
pcdh1g11
|
protocadherin 1 gamma 11 |
| chr25_+_20077225 | 2.53 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr8_+_50983551 | 2.52 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr18_+_28106139 | 2.51 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr8_+_554531 | 2.45 |
ENSDART00000193623
|
FO704758.2
|
|
| chr6_-_51101834 | 2.45 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr11_+_25112269 | 2.44 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr10_+_1638876 | 2.42 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr3_+_29600917 | 2.38 |
ENSDART00000048867
|
sstr3
|
somatostatin receptor 3 |
| chr11_-_41130239 | 2.38 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr12_-_7607114 | 2.37 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr13_+_36146415 | 2.37 |
ENSDART00000140301
|
TTC9
|
si:ch211-259k16.3 |
| chr3_+_33300522 | 2.37 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr17_-_43552894 | 2.36 |
ENSDART00000181226
ENSDART00000188125 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr21_+_9576176 | 2.36 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr3_+_30500968 | 2.35 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr2_+_55916911 | 2.29 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr1_+_6817292 | 2.29 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr16_+_12240605 | 2.29 |
ENSDART00000060056
|
tpi1b
|
triosephosphate isomerase 1b |
| chr4_-_13156971 | 2.28 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr20_-_45040916 | 2.27 |
ENSDART00000190001
|
klhl29
|
kelch-like family member 29 |
| chr7_-_19600181 | 2.24 |
ENSDART00000100757
|
oxa1l
|
oxidase (cytochrome c) assembly 1-like |
| chr8_+_52491436 | 2.18 |
ENSDART00000164298
|
si:ch1073-392o20.1
|
si:ch1073-392o20.1 |
| chr14_+_34547554 | 2.16 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr18_-_41232297 | 2.14 |
ENSDART00000036928
|
fbxo36a
|
F-box protein 36a |
| chr4_-_41269844 | 2.13 |
ENSDART00000186177
|
CR388165.2
|
|
| chr6_-_12135741 | 2.13 |
ENSDART00000155090
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr13_+_40034176 | 2.09 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr11_+_6882362 | 2.08 |
ENSDART00000144181
|
klhl26
|
kelch-like family member 26 |
| chr2_+_22363824 | 2.07 |
ENSDART00000163172
|
hs2st1a
|
heparan sulfate 2-O-sulfotransferase 1a |
| chr18_+_16192083 | 2.07 |
ENSDART00000133042
|
lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr14_-_48348973 | 2.06 |
ENSDART00000185822
|
CABZ01080056.1
|
|
| chr12_+_47280839 | 2.03 |
ENSDART00000187574
|
RYR2
|
ryanodine receptor 2 |
| chr8_+_53423408 | 2.02 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr20_+_27331008 | 2.02 |
ENSDART00000141486
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr3_+_32403758 | 2.02 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr19_-_1961024 | 2.02 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr1_+_52518176 | 1.99 |
ENSDART00000003278
|
tacr3l
|
tachykinin receptor 3-like |
| chr9_-_30346279 | 1.97 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
| chr23_+_2760573 | 1.97 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr7_+_50766094 | 1.96 |
ENSDART00000165037
|
si:ch73-380l10.2
|
si:ch73-380l10.2 |
| chr14_+_18785727 | 1.95 |
ENSDART00000184452
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr15_-_26887028 | 1.94 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr19_+_43563179 | 1.90 |
ENSDART00000151478
|
cd164l2
|
CD164 sialomucin-like 2 |
| chr18_-_8579907 | 1.85 |
ENSDART00000147284
|
FRMD4A
|
si:ch211-220f12.1 |
| chr7_-_72423666 | 1.84 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr17_-_45009782 | 1.79 |
ENSDART00000123971
|
fam161b
|
family with sequence similarity 161, member B |
| chr18_-_12295092 | 1.78 |
ENSDART00000033248
|
fam107b
|
family with sequence similarity 107, member B |
| chr12_+_48216662 | 1.75 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr2_-_24369087 | 1.75 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr9_-_7652792 | 1.75 |
ENSDART00000137957
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr10_-_20175118 | 1.74 |
ENSDART00000014417
|
htr7b
|
5-hydroxytryptamine (serotonin) receptor 7b |
| chr20_-_31075972 | 1.73 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr3_+_13929860 | 1.71 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr4_-_64871624 | 1.70 |
ENSDART00000124817
|
znf988
|
zinc finger protein 988 |
| chr24_-_25184553 | 1.69 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr22_+_10201826 | 1.69 |
ENSDART00000006513
ENSDART00000132641 |
pdhb
|
pyruvate dehydrogenase E1 beta subunit |
| chr9_-_21230999 | 1.69 |
ENSDART00000150160
ENSDART00000102147 |
pla1a
|
phospholipase A1 member A |
| chr9_-_43071519 | 1.68 |
ENSDART00000109099
|
ttn.2
|
titin, tandem duplicate 2 |
| chr9_-_25255490 | 1.67 |
ENSDART00000141502
|
htr2aa
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate a |
| chr7_+_26709251 | 1.67 |
ENSDART00000149426
ENSDART00000010323 |
cd82a
|
CD82 molecule a |
| chr21_+_30084823 | 1.66 |
ENSDART00000154573
|
prob1
|
proline-rich basic protein 1 |
| chr17_-_24564674 | 1.63 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr16_-_21047872 | 1.63 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr3_+_25508247 | 1.61 |
ENSDART00000039448
|
mchr1b
|
melanin-concentrating hormone receptor 1b |
| chr5_+_53482597 | 1.60 |
ENSDART00000180333
|
BX323994.1
|
|
| chr24_-_34034931 | 1.57 |
ENSDART00000169227
|
asic1c
|
acid-sensing (proton-gated) ion channel 1c |
| chr17_+_20204122 | 1.55 |
ENSDART00000078672
|
gnrh3
|
gonadotropin-releasing hormone 3 |
| chr6_-_33685325 | 1.54 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr23_+_37185247 | 1.52 |
ENSDART00000146269
|
vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr14_+_1687357 | 1.52 |
ENSDART00000174675
|
hrh2a
|
histamine receptor H2a |
| chr16_-_28727763 | 1.47 |
ENSDART00000149575
|
dcst1
|
DC-STAMP domain containing 1 |
| chr1_+_34203817 | 1.47 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr10_-_41664427 | 1.47 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr20_+_48754045 | 1.46 |
ENSDART00000062578
|
zgc:110348
|
zgc:110348 |
| chr12_-_3077395 | 1.46 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr23_-_28347039 | 1.45 |
ENSDART00000145072
|
neurod4
|
neuronal differentiation 4 |
| chr6_+_27339962 | 1.45 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr17_+_37932706 | 1.43 |
ENSDART00000075941
|
plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr5_-_35888499 | 1.43 |
ENSDART00000193932
|
rxfp2l
|
relaxin/insulin-like family peptide receptor 2, like |
| chr13_+_10023256 | 1.42 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr5_-_24642026 | 1.42 |
ENSDART00000139522
|
si:ch211-106a19.1
|
si:ch211-106a19.1 |
| chr21_+_10773522 | 1.39 |
ENSDART00000143257
|
znf532
|
zinc finger protein 532 |
| chr4_-_39111612 | 1.39 |
ENSDART00000150394
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr14_+_31721171 | 1.38 |
ENSDART00000173034
|
adgrg4a
|
adhesion G protein-coupled receptor G4a |
| chr22_-_32392252 | 1.37 |
ENSDART00000148681
ENSDART00000137945 |
pcbp4
|
poly(rC) binding protein 4 |
| chr8_+_21406769 | 1.34 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr7_+_34290051 | 1.33 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr3_+_30501135 | 1.33 |
ENSDART00000165869
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr2_+_12255568 | 1.32 |
ENSDART00000184164
ENSDART00000013454 |
prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr23_+_19675780 | 1.32 |
ENSDART00000124335
|
smim4
|
small integral membrane protein 4 |
| chr7_-_35516251 | 1.32 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr7_+_13609457 | 1.31 |
ENSDART00000172857
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr22_+_19478140 | 1.31 |
ENSDART00000135291
ENSDART00000136576 |
si:dkey-78l4.8
|
si:dkey-78l4.8 |
| chr20_+_46183505 | 1.30 |
ENSDART00000060799
|
taar13b
|
trace amine associated receptor 13b |
| chr7_-_11020900 | 1.29 |
ENSDART00000182976
|
BX470137.1
|
|
| chr6_-_36182115 | 1.28 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr5_+_53824959 | 1.28 |
ENSDART00000169565
|
sat2b
|
spermidine/spermine N1-acetyltransferase family member 2b |
| chr10_+_17592273 | 1.27 |
ENSDART00000141221
|
KCNN2
|
si:dkey-76p7.5 |
| chr19_+_8985230 | 1.27 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr4_+_22343093 | 1.26 |
ENSDART00000023588
|
guca1a
|
guanylate cyclase activator 1A |
| chr5_-_9073433 | 1.26 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr10_-_40448736 | 1.25 |
ENSDART00000137644
ENSDART00000168190 |
taar20p
|
trace amine associated receptor 20p |
| chr14_+_4276394 | 1.23 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr10_-_19006579 | 1.22 |
ENSDART00000141764
|
adra1ab
|
adrenoceptor alpha 1Ab |
| chr22_+_2254972 | 1.21 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr19_-_17385548 | 1.21 |
ENSDART00000162383
|
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr2_+_10642047 | 1.20 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr3_-_33029426 | 1.19 |
ENSDART00000181287
|
CT583642.1
|
|
| chr16_+_43077909 | 1.19 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of maff+mafga+mafgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 11.4 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 1.2 | 5.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 1.1 | 3.2 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) neuron projection maintenance(GO:1990535) |
| 1.0 | 4.0 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 1.0 | 5.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.0 | 5.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.9 | 4.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.7 | 7.4 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.7 | 3.7 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.6 | 3.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.6 | 2.3 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.6 | 5.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.6 | 2.2 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.6 | 2.8 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.5 | 7.7 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.5 | 8.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.5 | 2.5 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.5 | 2.9 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.4 | 1.2 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.4 | 2.0 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.4 | 6.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.4 | 2.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 3.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 1.5 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.3 | 0.9 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.3 | 6.0 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.3 | 6.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 2.0 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.3 | 1.7 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.3 | 4.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.3 | 1.4 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.3 | 1.6 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.2 | 8.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 0.9 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 8.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 0.9 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.2 | 0.9 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.2 | 0.9 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.2 | 2.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 1.5 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.2 | 3.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.2 | 0.8 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) dGDP metabolic process(GO:0046066) |
| 0.2 | 2.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 3.5 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.2 | 1.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 2.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 2.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 2.3 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.2 | 1.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.2 | 2.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 3.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 0.7 | GO:0033605 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.2 | 1.2 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.2 | 0.8 | GO:0045822 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.2 | 1.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.6 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.1 | 7.1 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 13.3 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.1 | 0.9 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 1.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.5 | GO:0070285 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) pigment cell development(GO:0070285) |
| 0.1 | 2.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 6.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 1.4 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 3.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.7 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 4.7 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 5.6 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.3 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 0.6 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 1.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 2.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.6 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 2.7 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.1 | 4.6 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.1 | 0.4 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.9 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 3.2 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.6 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 3.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 2.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 2.1 | GO:0042310 | vasoconstriction(GO:0042310) |
| 0.1 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.9 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 1.4 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.1 | 4.1 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 3.8 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 4.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 3.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 1.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.5 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.4 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.5 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 1.3 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.7 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 6.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.1 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.4 | GO:0001966 | thigmotaxis(GO:0001966) myotome development(GO:0061055) |
| 0.0 | 0.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 3.8 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.5 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.9 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 1.3 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 0.4 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.3 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 2.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 1.1 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.6 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.1 | GO:0039022 | pronephric duct development(GO:0039022) nephric duct development(GO:0072176) |
| 0.0 | 0.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.4 | GO:0050769 | positive regulation of neurogenesis(GO:0050769) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.7 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.4 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.3 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 2.2 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 2.8 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.8 | 4.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.8 | 4.6 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.5 | 1.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.5 | 6.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.5 | 8.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 2.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.3 | 4.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 2.9 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.3 | 1.7 | GO:0000801 | central element(GO:0000801) |
| 0.3 | 1.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 2.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 3.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 3.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 9.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 2.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 5.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 10.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 3.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 6.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 4.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 5.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 3.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 1.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 2.8 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 2.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 2.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.9 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.2 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.0 | 2.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 8.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 5.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 12.4 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 9.0 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 1.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.7 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.3 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 18.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.6 | GO:0005929 | cilium(GO:0005929) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 1.7 | 5.0 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 1.2 | 4.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.0 | 3.1 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 1.0 | 2.9 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.9 | 8.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.6 | 6.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 2.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.5 | 1.6 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.5 | 1.5 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.4 | 7.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.4 | 1.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 5.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.4 | 5.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.4 | 1.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 3.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 3.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.3 | 1.7 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.3 | 8.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.3 | 6.0 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.3 | 1.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 3.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 0.9 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.3 | 2.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.3 | 2.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.3 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 1.0 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.2 | 2.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.2 | 6.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 2.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 2.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 2.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 5.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.6 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 2.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 3.7 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 3.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 1.0 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.2 | 7.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 1.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 1.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 2.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 2.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 2.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 5.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 3.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.6 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 5.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 5.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.7 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.6 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.9 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 2.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 6.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 3.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.4 | GO:0099589 | serotonin-gated cation channel activity(GO:0022850) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 2.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 2.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.0 | 1.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 2.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.8 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 1.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 2.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.8 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 2.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 7.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 13.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0035255 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 17.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 3.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 3.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 3.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 3.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.9 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.6 | 6.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 4.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 3.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 7.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |