Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
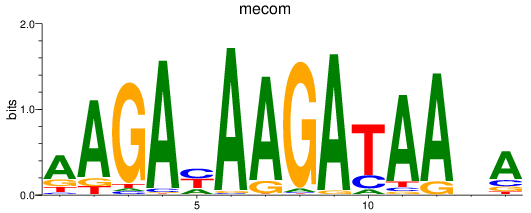
Results for mecom
Z-value: 0.71
Transcription factors associated with mecom
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mecom
|
ENSDARG00000060808 | MDS1 and EVI1 complex locus |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mecom | dr11_v1_chr15_-_35410860_35410860 | -0.42 | 2.9e-05 | Click! |
Activity profile of mecom motif
Sorted Z-values of mecom motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61203203 | 10.18 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr15_+_22311803 | 5.50 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr17_-_125091 | 5.32 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr3_-_32817274 | 5.20 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr1_+_31110817 | 4.82 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr14_+_35691889 | 4.75 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr1_-_45553602 | 4.33 |
ENSDART00000143664
|
grin2bb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b |
| chr3_+_32443395 | 4.08 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr6_-_40697585 | 3.94 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr24_-_15648636 | 3.91 |
ENSDART00000136200
|
cbln2b
|
cerebellin 2b precursor |
| chr10_-_31782616 | 3.77 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr2_-_44283554 | 3.61 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr22_-_29242347 | 3.51 |
ENSDART00000040761
|
pvalb7
|
parvalbumin 7 |
| chr15_-_23647078 | 3.23 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr1_+_9071324 | 3.21 |
ENSDART00000184194
|
BX004779.2
|
|
| chr18_-_21271373 | 3.05 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr23_+_44745317 | 3.01 |
ENSDART00000165654
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr11_-_37509001 | 2.90 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr10_+_22012389 | 2.87 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr2_+_2563192 | 2.80 |
ENSDART00000055713
|
CABZ01033000.1
|
|
| chr22_+_11535131 | 2.73 |
ENSDART00000113930
|
npb
|
neuropeptide B |
| chr19_+_16222618 | 2.72 |
ENSDART00000137189
ENSDART00000169246 ENSDART00000190583 ENSDART00000189521 |
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| chr20_-_34801181 | 2.68 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr16_-_52879741 | 2.63 |
ENSDART00000166470
|
TPPP
|
tubulin polymerization promoting protein |
| chr7_-_38612230 | 2.55 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr11_+_39672874 | 2.53 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr6_-_50203682 | 2.52 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr4_+_26628822 | 2.51 |
ENSDART00000191030
ENSDART00000186113 ENSDART00000186764 ENSDART00000165158 |
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr9_+_29585943 | 2.47 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr9_+_13714379 | 2.36 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr21_+_21326038 | 2.32 |
ENSDART00000089898
|
qpctlb
|
glutaminyl-peptide cyclotransferase-like b |
| chr12_-_6551681 | 2.20 |
ENSDART00000145413
|
si:ch211-253p2.2
|
si:ch211-253p2.2 |
| chr21_+_5209716 | 2.19 |
ENSDART00000102539
ENSDART00000053148 ENSDART00000102536 |
st8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr11_+_35364445 | 2.17 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr15_+_33991928 | 2.16 |
ENSDART00000170177
|
vwde
|
von Willebrand factor D and EGF domains |
| chr2_+_29976419 | 2.10 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr9_-_9348058 | 2.09 |
ENSDART00000132257
|
zgc:113337
|
zgc:113337 |
| chr3_+_37824268 | 2.06 |
ENSDART00000137038
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr17_+_33495194 | 2.04 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr9_+_31534601 | 2.04 |
ENSDART00000133094
|
si:ch211-168k14.2
|
si:ch211-168k14.2 |
| chr14_-_2187202 | 2.03 |
ENSDART00000160205
|
pcdh2ab12
|
protocadherin 2 alpha b 12 |
| chr13_-_11644806 | 2.01 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr16_+_12836143 | 1.94 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr22_+_33362552 | 1.91 |
ENSDART00000101580
|
nicn1
|
nicolin 1 |
| chr16_+_30959423 | 1.89 |
ENSDART00000136824
|
gstk2
|
glutathione S-transferase kappa 2 |
| chr11_-_23322182 | 1.86 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr22_-_15385442 | 1.86 |
ENSDART00000090975
|
tmem264
|
transmembrane protein 264 |
| chr16_+_7662609 | 1.85 |
ENSDART00000184895
ENSDART00000149404 ENSDART00000081418 ENSDART00000081422 |
bves
|
blood vessel epicardial substance |
| chr13_+_10232695 | 1.83 |
ENSDART00000080805
|
six2a
|
SIX homeobox 2a |
| chr9_+_1138323 | 1.83 |
ENSDART00000190352
ENSDART00000190387 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr22_+_8800432 | 1.82 |
ENSDART00000139940
|
si:dkey-182g1.6
|
si:dkey-182g1.6 |
| chr7_+_10351038 | 1.81 |
ENSDART00000173256
|
si:cabz01029535.1
|
si:cabz01029535.1 |
| chr19_+_17259912 | 1.80 |
ENSDART00000078951
|
MANEAL
|
si:ch211-30b16.2 |
| chr6_-_6423885 | 1.79 |
ENSDART00000092257
|
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr1_+_37752171 | 1.78 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr25_+_25438165 | 1.74 |
ENSDART00000161369
|
LRRC56
|
si:ch211-103e16.5 |
| chr9_+_38983895 | 1.69 |
ENSDART00000144893
|
map2
|
microtubule-associated protein 2 |
| chr16_+_14029283 | 1.68 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr9_+_10014817 | 1.67 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr14_+_22591624 | 1.65 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr17_-_42213822 | 1.65 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr15_-_8517555 | 1.63 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr21_-_12119711 | 1.61 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr1_+_33383971 | 1.61 |
ENSDART00000150043
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr5_-_10946232 | 1.60 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr7_-_38340674 | 1.59 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr15_+_16908085 | 1.58 |
ENSDART00000186870
|
ypel2b
|
yippee-like 2b |
| chr6_-_39765546 | 1.56 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr21_+_15704556 | 1.56 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr6_-_46861676 | 1.56 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr24_-_30096666 | 1.54 |
ENSDART00000183285
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr17_+_26981132 | 1.52 |
ENSDART00000156672
|
si:dkey-221l4.11
|
si:dkey-221l4.11 |
| chr3_-_33437796 | 1.52 |
ENSDART00000075499
|
si:dkey-283b1.7
|
si:dkey-283b1.7 |
| chr18_-_6633984 | 1.50 |
ENSDART00000185241
|
tnni1c
|
troponin I, skeletal, slow c |
| chr16_+_36126310 | 1.50 |
ENSDART00000166040
ENSDART00000189802 |
sh3bp5b
|
SH3-domain binding protein 5b (BTK-associated) |
| chr9_+_48455909 | 1.50 |
ENSDART00000167243
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr22_+_10752787 | 1.50 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr4_-_27398385 | 1.50 |
ENSDART00000142117
ENSDART00000150553 ENSDART00000182746 |
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr24_+_26276805 | 1.49 |
ENSDART00000089749
|
adipoqa
|
adiponectin, C1Q and collagen domain containing, a |
| chr2_-_1188707 | 1.47 |
ENSDART00000066378
|
CABZ01084564.1
|
|
| chr13_-_39830999 | 1.47 |
ENSDART00000115089
|
zgc:171482
|
zgc:171482 |
| chr23_-_30781875 | 1.43 |
ENSDART00000114628
ENSDART00000180949 ENSDART00000191313 |
myt1a
|
myelin transcription factor 1a |
| chr24_+_41915878 | 1.41 |
ENSDART00000171523
|
TMEM200C
|
transmembrane protein 200C |
| chr11_-_3343463 | 1.39 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr10_-_15672862 | 1.39 |
ENSDART00000109231
|
mamdc2b
|
MAM domain containing 2b |
| chr8_+_48801402 | 1.38 |
ENSDART00000098363
|
tprg1l
|
tumor protein p63 regulated 1-like |
| chr5_-_31926906 | 1.37 |
ENSDART00000187340
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr9_+_10014514 | 1.35 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr2_+_38554260 | 1.33 |
ENSDART00000171527
|
cdh24b
|
cadherin 24, type 2b |
| chr22_+_10752511 | 1.33 |
ENSDART00000081188
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr17_+_29348013 | 1.31 |
ENSDART00000188885
ENSDART00000192005 ENSDART00000190484 ENSDART00000188565 |
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr22_+_8799855 | 1.30 |
ENSDART00000078235
|
si:dkey-182g1.6
|
si:dkey-182g1.6 |
| chr5_+_44190974 | 1.29 |
ENSDART00000182634
ENSDART00000190626 |
TMEM8B
|
si:dkey-84j12.1 |
| chr13_+_12174937 | 1.29 |
ENSDART00000171683
|
gabrg1
|
gamma-aminobutyric acid type A receptor gamma1 subunit |
| chr5_-_5147041 | 1.29 |
ENSDART00000180236
|
lmx1ba
|
LIM homeobox transcription factor 1, beta a |
| chr22_-_37565348 | 1.28 |
ENSDART00000149482
ENSDART00000104478 |
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr23_+_35969228 | 1.26 |
ENSDART00000179992
ENSDART00000184107 |
hoxc10a
|
homeobox C10a |
| chr10_-_30785501 | 1.25 |
ENSDART00000020054
|
opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr10_-_5581487 | 1.25 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr24_+_25471196 | 1.25 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr12_-_29301022 | 1.23 |
ENSDART00000187826
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr19_+_42693855 | 1.23 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr17_+_13763425 | 1.23 |
ENSDART00000105175
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr2_-_1548330 | 1.22 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr25_-_16782394 | 1.22 |
ENSDART00000019413
|
ndufa9a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a |
| chr7_-_17028015 | 1.21 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr9_-_1986014 | 1.21 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr2_-_10188598 | 1.21 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr1_+_38758261 | 1.20 |
ENSDART00000182756
|
wdr17
|
WD repeat domain 17 |
| chr21_+_41743493 | 1.20 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr13_+_29771463 | 1.19 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr9_+_2002701 | 1.19 |
ENSDART00000082329
|
evx2
|
even-skipped homeobox 2 |
| chr2_-_3678029 | 1.17 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr7_+_21887307 | 1.16 |
ENSDART00000052871
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr1_+_38758445 | 1.16 |
ENSDART00000136300
|
wdr17
|
WD repeat domain 17 |
| chr11_+_42633376 | 1.15 |
ENSDART00000076711
|
appl1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr4_-_12104421 | 1.11 |
ENSDART00000139561
|
mrps33
|
mitochondrial ribosomal protein S33 |
| chr21_-_43398457 | 1.10 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr1_+_34181581 | 1.10 |
ENSDART00000146042
|
epha6
|
eph receptor A6 |
| chr12_+_38774860 | 1.09 |
ENSDART00000130371
|
kif19
|
kinesin family member 19 |
| chr14_+_33722950 | 1.07 |
ENSDART00000075312
|
apln
|
apelin |
| chr6_-_29195642 | 1.06 |
ENSDART00000078625
|
dpt
|
dermatopontin |
| chr24_-_31843173 | 1.06 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr9_+_1137430 | 1.04 |
ENSDART00000187062
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr25_+_25438322 | 1.03 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr11_-_37359416 | 1.01 |
ENSDART00000159184
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr20_+_38322444 | 1.01 |
ENSDART00000161741
ENSDART00000132241 ENSDART00000148936 ENSDART00000149623 |
stum
|
stum, mechanosensory transduction mediator homolog |
| chr19_+_19511394 | 0.99 |
ENSDART00000172410
|
jazf1a
|
JAZF zinc finger 1a |
| chr18_+_16330025 | 0.99 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr25_-_12935065 | 0.98 |
ENSDART00000167362
|
ccl39.7
|
chemokine (C-C motif) ligand 39, duplicate 7 |
| chr16_+_19014886 | 0.98 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr2_+_15776649 | 0.98 |
ENSDART00000156535
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr3_-_18274691 | 0.96 |
ENSDART00000161140
|
BX649434.3
|
|
| chr14_-_32089117 | 0.96 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr3_+_1637358 | 0.94 |
ENSDART00000180266
|
CR394546.5
|
|
| chr25_+_33002963 | 0.93 |
ENSDART00000187366
|
CABZ01046980.1
|
|
| chr2_-_51290607 | 0.90 |
ENSDART00000171693
|
si:ch211-215e19.3
|
si:ch211-215e19.3 |
| chr4_-_8902406 | 0.88 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr6_-_25181012 | 0.88 |
ENSDART00000161585
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr15_+_19646902 | 0.88 |
ENSDART00000179822
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr13_+_30172645 | 0.87 |
ENSDART00000137114
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr3_+_32789605 | 0.86 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr14_-_30141162 | 0.85 |
ENSDART00000053916
|
mtnr1ab
|
melatonin receptor 1A b |
| chr25_-_12935424 | 0.84 |
ENSDART00000160217
|
ccl39.7
|
chemokine (C-C motif) ligand 39, duplicate 7 |
| chr15_+_5028608 | 0.84 |
ENSDART00000092809
|
abcg1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr9_-_18742704 | 0.82 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr24_-_26304386 | 0.81 |
ENSDART00000175416
|
otos
|
otospiralin |
| chr16_+_10972733 | 0.79 |
ENSDART00000049323
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr4_+_54618332 | 0.79 |
ENSDART00000171824
|
si:ch211-227e10.2
|
si:ch211-227e10.2 |
| chr20_+_1996202 | 0.78 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr10_+_34394454 | 0.78 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr15_+_28119704 | 0.78 |
ENSDART00000184290
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr18_-_28938912 | 0.76 |
ENSDART00000136201
|
si:ch211-174j14.2
|
si:ch211-174j14.2 |
| chr10_+_21899753 | 0.76 |
ENSDART00000080155
|
hrh2b
|
histamine receptor H2b |
| chr6_-_31806345 | 0.74 |
ENSDART00000149369
|
si:dkey-209n16.2
|
si:dkey-209n16.2 |
| chr6_+_8176486 | 0.74 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr23_+_20110086 | 0.74 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr12_+_23912074 | 0.73 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr12_+_2428247 | 0.73 |
ENSDART00000152529
|
lrrc18b
|
leucine rich repeat containing 18b |
| chr23_+_30835589 | 0.72 |
ENSDART00000154145
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr1_-_36771712 | 0.72 |
ENSDART00000148386
|
prmt9
|
protein arginine methyltransferase 9 |
| chr7_+_38395197 | 0.71 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr1_-_33351780 | 0.71 |
ENSDART00000178996
|
gyg2
|
glycogenin 2 |
| chr6_-_29051773 | 0.70 |
ENSDART00000190508
ENSDART00000180191 ENSDART00000111682 |
evi5b
|
ecotropic viral integration site 5b |
| chr15_-_8517376 | 0.69 |
ENSDART00000186289
|
npas1
|
neuronal PAS domain protein 1 |
| chr18_+_16246806 | 0.68 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr5_+_19165949 | 0.68 |
ENSDART00000140710
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr13_-_39830816 | 0.67 |
ENSDART00000138981
|
zgc:171482
|
zgc:171482 |
| chr3_+_24511959 | 0.66 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr21_+_30043054 | 0.65 |
ENSDART00000065448
|
fabp6
|
fatty acid binding protein 6, ileal (gastrotropin) |
| chr4_-_39110934 | 0.65 |
ENSDART00000185041
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr19_-_36401282 | 0.65 |
ENSDART00000193856
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr24_-_5889878 | 0.65 |
ENSDART00000077951
|
pcolce2b
|
procollagen C-endopeptidase enhancer 2b |
| chr24_-_26460853 | 0.64 |
ENSDART00000078628
|
slc7a14b
|
solute carrier family 7, member 14b |
| chr6_-_10964083 | 0.64 |
ENSDART00000181583
|
notum2
|
notum pectinacetylesterase 2 |
| chr11_-_41130239 | 0.64 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr20_+_48803248 | 0.64 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr19_+_27342479 | 0.63 |
ENSDART00000184687
|
ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr6_+_43390387 | 0.63 |
ENSDART00000122793
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr12_-_32426685 | 0.63 |
ENSDART00000105574
|
enpp7.2
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 2 |
| chr3_-_147368 | 0.60 |
ENSDART00000159073
|
CR391944.1
|
|
| chr7_-_8374950 | 0.59 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr6_+_46259950 | 0.59 |
ENSDART00000032326
|
zgc:162324
|
zgc:162324 |
| chr5_-_18046053 | 0.59 |
ENSDART00000144898
|
rnf215
|
ring finger protein 215 |
| chr6_-_25180438 | 0.58 |
ENSDART00000159696
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr2_+_17540720 | 0.58 |
ENSDART00000114638
|
pimr198
|
Pim proto-oncogene, serine/threonine kinase, related 198 |
| chr15_+_6774994 | 0.58 |
ENSDART00000170011
|
si:ch1073-228b5.2
|
si:ch1073-228b5.2 |
| chr10_-_22854758 | 0.57 |
ENSDART00000079449
|
and3
|
actinodin3 |
| chr7_+_5541747 | 0.56 |
ENSDART00000144876
|
si:dkeyp-67a8.4
|
si:dkeyp-67a8.4 |
| chr23_+_19213472 | 0.56 |
ENSDART00000185985
|
apobec2b
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2b |
| chr24_+_31348595 | 0.55 |
ENSDART00000160140
ENSDART00000158661 |
cremb
|
cAMP responsive element modulator b |
| chr18_-_6634424 | 0.54 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr22_-_17256573 | 0.53 |
ENSDART00000136119
ENSDART00000062891 |
nphs2
|
nephrosis 2, idiopathic, steroid-resistant (podocin) |
| chr13_+_18514788 | 0.53 |
ENSDART00000124036
|
tlr4ba
|
toll-like receptor 4b, duplicate a |
| chr5_-_5147220 | 0.53 |
ENSDART00000187026
ENSDART00000162334 |
lmx1ba
|
LIM homeobox transcription factor 1, beta a |
| chr25_-_1235457 | 0.52 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr25_-_4235037 | 0.52 |
ENSDART00000093003
|
syt7a
|
synaptotagmin VIIa |
| chr23_-_20154422 | 0.52 |
ENSDART00000132025
|
usp19
|
ubiquitin specific peptidase 19 |
| chr1_+_22002649 | 0.52 |
ENSDART00000141317
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr14_-_47963115 | 0.51 |
ENSDART00000003826
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mecom
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.5 | 1.6 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.5 | 1.6 | GO:0042942 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.4 | 1.2 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.4 | 1.2 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.4 | 2.0 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.4 | 1.9 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.3 | 2.9 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.3 | 4.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.3 | 2.9 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.3 | 1.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 3.2 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 4.3 | GO:0099565 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.2 | 0.8 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.2 | 0.8 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.2 | 1.5 | GO:0070293 | renal absorption(GO:0070293) |
| 0.2 | 0.6 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.2 | 1.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.2 | 0.5 | GO:0071312 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.2 | 3.0 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 3.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 1.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 1.8 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 1.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 1.6 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 1.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.5 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.7 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 1.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.4 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 2.7 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 0.8 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 2.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.3 | GO:0060078 | regulation of postsynaptic membrane potential(GO:0060078) |
| 0.1 | 1.0 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.1 | 1.9 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.1 | 2.0 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 4.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.5 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.1 | 1.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 3.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.2 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 2.7 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.1 | 0.2 | GO:0046436 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 2.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 1.9 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 1.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.3 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.2 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.6 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.9 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.7 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 1.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.7 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 2.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.4 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.5 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 1.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 1.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.3 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.6 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 2.2 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.6 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.6 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.5 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 2.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 5.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 1.3 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.3 | 3.0 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 2.9 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.3 | 4.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 1.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 3.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 1.7 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 0.5 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 4.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 2.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.3 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.5 | 4.7 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.4 | 2.9 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.4 | 1.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.3 | 3.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 2.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.3 | 2.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 1.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 3.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 3.9 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 4.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 0.8 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.2 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 1.9 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.2 | 2.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 0.5 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.2 | 2.0 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.2 | 0.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 3.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.4 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.7 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 4.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 3.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.7 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.8 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 1.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.5 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 0.5 | GO:0052743 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 1.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0015250 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 1.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 2.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.5 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.5 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 4.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 4.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 3.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 2.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |