Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
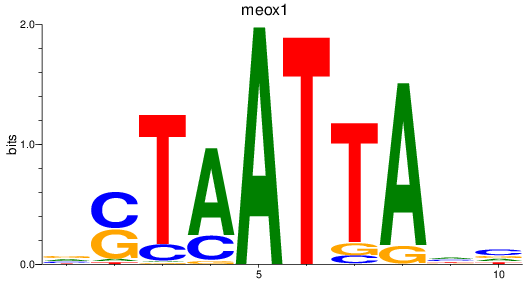
Results for meox1
Z-value: 0.82
Transcription factors associated with meox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox1
|
ENSDARG00000007891 | mesenchyme homeobox 1 |
|
meox1
|
ENSDARG00000115382 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox1 | dr11_v1_chr12_+_27462225_27462225 | 0.15 | 1.5e-01 | Click! |
Activity profile of meox1 motif
Sorted Z-values of meox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_1185772 | 11.24 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr16_+_5774977 | 10.62 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr19_-_32641725 | 9.12 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr11_+_25129013 | 9.04 |
ENSDART00000132879
|
ndrg3a
|
ndrg family member 3a |
| chr9_-_44295071 | 8.78 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr20_-_9462433 | 8.60 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr7_+_30787903 | 8.36 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr19_-_5103141 | 8.17 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr23_+_28731379 | 7.35 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr4_+_21129752 | 7.34 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr9_+_38372216 | 6.91 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr20_-_9436521 | 6.75 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr16_+_46111849 | 6.19 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr24_+_16547035 | 6.01 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr24_-_41320037 | 5.93 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr22_+_17205608 | 5.84 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr2_+_50608099 | 5.75 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr19_-_5103313 | 5.66 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr1_+_40023640 | 5.60 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr1_-_50859053 | 5.58 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr20_-_28800999 | 5.35 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr22_+_17828267 | 5.31 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr12_-_33972798 | 5.30 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr6_-_35472923 | 5.28 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr5_+_64732270 | 4.92 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr15_+_15856178 | 4.89 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr13_-_29420885 | 4.81 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr13_+_33117528 | 4.55 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr5_+_38263240 | 4.45 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr9_-_43538328 | 4.42 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr19_-_20113696 | 4.42 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr1_+_16397063 | 4.37 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr23_+_40460333 | 4.30 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr9_+_11532025 | 4.24 |
ENSDART00000109037
|
cdk5r2b
|
cyclin-dependent kinase 5, regulatory subunit 2b (p39) |
| chr4_+_11384891 | 4.20 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr18_+_9637744 | 4.20 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr17_-_28198099 | 4.11 |
ENSDART00000156143
|
htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr3_+_34919810 | 4.10 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr20_-_20931197 | 4.05 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr20_-_29864390 | 3.85 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr15_-_9272328 | 3.77 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr21_-_25604603 | 3.70 |
ENSDART00000133134
ENSDART00000139460 |
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr7_-_25895189 | 3.68 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr7_-_28148310 | 3.61 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr6_-_6487876 | 3.60 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr14_+_45406299 | 3.58 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr20_-_35040041 | 3.57 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr7_+_23292133 | 3.44 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr16_+_17389116 | 3.39 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr1_-_10647484 | 3.38 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr10_-_41400049 | 3.37 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr11_-_44801968 | 3.32 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr4_-_17629444 | 3.27 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr1_-_10647307 | 3.25 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr18_+_11506561 | 3.24 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr3_+_32443395 | 3.22 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr17_+_45454943 | 3.19 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr6_+_28208973 | 3.11 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr25_-_27722614 | 3.08 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr22_-_11137268 | 3.06 |
ENSDART00000178882
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr7_-_30174882 | 3.01 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr6_+_21001264 | 2.98 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr10_-_34772211 | 2.90 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr20_-_20930926 | 2.85 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr2_-_16380283 | 2.84 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr3_+_33341640 | 2.81 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr22_+_5176255 | 2.75 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr11_+_28476298 | 2.69 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr3_+_22442445 | 2.68 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr17_+_11675362 | 2.64 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr9_-_38021889 | 2.64 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr6_-_44044385 | 2.64 |
ENSDART00000075497
|
rybpb
|
RING1 and YY1 binding protein b |
| chr11_-_36341028 | 2.62 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr11_+_38280454 | 2.53 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr9_+_17983463 | 2.51 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr21_-_27185915 | 2.51 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr21_+_26726936 | 2.49 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr5_+_30635309 | 2.48 |
ENSDART00000183769
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr10_+_29698467 | 2.47 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr2_+_6253246 | 2.45 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr11_-_23687158 | 2.45 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr18_+_9615147 | 2.43 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr20_-_23226453 | 2.41 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr18_-_40708537 | 2.40 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr24_+_36204028 | 2.36 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr6_-_12275836 | 2.36 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr18_+_1154189 | 2.28 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr15_+_21276735 | 2.26 |
ENSDART00000111213
|
ubash3bb
|
ubiquitin associated and SH3 domain containing Bb |
| chr3_-_46817499 | 2.26 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr7_-_59165640 | 2.25 |
ENSDART00000170853
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr18_+_14529005 | 2.24 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr16_+_24684107 | 2.15 |
ENSDART00000183920
|
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr15_-_18432673 | 2.11 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr11_+_36231248 | 2.11 |
ENSDART00000131104
|
GPR62 (1 of many)
|
si:ch211-213o11.11 |
| chr19_+_47476908 | 2.05 |
ENSDART00000157886
|
zgc:114119
|
zgc:114119 |
| chr11_-_1550709 | 1.99 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr25_+_5035343 | 1.99 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr6_+_39232245 | 1.95 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr2_-_23172708 | 1.88 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr2_-_33645411 | 1.88 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr24_+_22485710 | 1.85 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr5_+_20147830 | 1.83 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr3_-_61162750 | 1.79 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr24_+_12133814 | 1.79 |
ENSDART00000158562
ENSDART00000159029 ENSDART00000168248 |
lztfl1
|
leucine zipper transcription factor-like 1 |
| chr7_+_20110336 | 1.79 |
ENSDART00000179395
|
zgc:114045
|
zgc:114045 |
| chr22_-_10459880 | 1.78 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr20_-_46362606 | 1.77 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr12_-_33357655 | 1.76 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr11_-_6974022 | 1.76 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr22_-_3564563 | 1.75 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr20_-_27225876 | 1.75 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr2_+_1714640 | 1.75 |
ENSDART00000086761
ENSDART00000111613 |
adgrl2b.1
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr23_+_31913292 | 1.72 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr6_-_55585423 | 1.71 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr20_+_40457599 | 1.70 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr20_-_14925281 | 1.69 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr4_-_16124417 | 1.68 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr7_-_27685365 | 1.67 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr25_-_27722309 | 1.62 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr1_-_44701313 | 1.62 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr6_-_12172424 | 1.62 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr11_+_24620742 | 1.60 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr23_-_36446307 | 1.60 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr11_+_30244356 | 1.58 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr3_+_1637358 | 1.58 |
ENSDART00000180266
|
CR394546.5
|
|
| chr24_+_40860320 | 1.58 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr17_+_21902058 | 1.56 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr17_-_31695217 | 1.54 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr20_+_26966725 | 1.52 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr19_+_22062202 | 1.52 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr11_-_472547 | 1.47 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr23_-_10175898 | 1.46 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr20_-_38787341 | 1.43 |
ENSDART00000136771
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr22_+_5176693 | 1.42 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr12_-_28349026 | 1.41 |
ENSDART00000183768
ENSDART00000152998 |
zgc:195081
|
zgc:195081 |
| chr14_+_25817628 | 1.40 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr25_+_29160102 | 1.39 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr15_-_1038193 | 1.37 |
ENSDART00000159462
|
si:dkey-77f5.3
|
si:dkey-77f5.3 |
| chr1_+_17676745 | 1.36 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr18_+_8402129 | 1.34 |
ENSDART00000081154
ENSDART00000171974 |
prpf18
|
PRP18 pre-mRNA processing factor 18 homolog (yeast) |
| chr11_-_34219211 | 1.32 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr2_+_32826235 | 1.30 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr14_-_30587814 | 1.29 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr9_-_35633827 | 1.28 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr20_-_14924858 | 1.28 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr16_+_43347966 | 1.27 |
ENSDART00000171308
|
zmp:0000000930
|
zmp:0000000930 |
| chr3_+_45364849 | 1.27 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr2_-_11258547 | 1.26 |
ENSDART00000165803
ENSDART00000193817 |
slc44a5a
|
solute carrier family 44, member 5a |
| chr11_-_2131280 | 1.24 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr20_+_29209615 | 1.23 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr15_+_29140126 | 1.23 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr3_-_60827402 | 1.22 |
ENSDART00000053494
|
anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr14_+_34971554 | 1.18 |
ENSDART00000184271
|
rnf145a
|
ring finger protein 145a |
| chr5_+_19314574 | 1.17 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr6_-_40713183 | 1.16 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr18_-_2433011 | 1.16 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr18_-_16590056 | 1.16 |
ENSDART00000143744
|
mgat4c
|
mgat4 family, member C |
| chr3_-_26806032 | 1.13 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr23_-_31913231 | 1.11 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr19_+_15521997 | 1.11 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr3_+_45365098 | 1.10 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr8_-_23780334 | 1.09 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr4_-_28158335 | 1.08 |
ENSDART00000134605
|
gramd4a
|
GRAM domain containing 4a |
| chr15_-_22074315 | 1.07 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr22_-_13350240 | 1.06 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr7_+_7511914 | 1.06 |
ENSDART00000172848
|
clcn3
|
chloride channel 3 |
| chr9_+_43799829 | 1.05 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr20_-_38787047 | 1.04 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr15_-_590787 | 1.02 |
ENSDART00000189367
|
si:ch73-144d13.5
|
si:ch73-144d13.5 |
| chr21_-_3770636 | 1.01 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr5_-_68093169 | 1.00 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr10_+_16036246 | 0.98 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr24_-_39518599 | 0.95 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr15_+_33865312 | 0.95 |
ENSDART00000166141
|
tekt1
|
tektin 1 |
| chr15_+_44366556 | 0.95 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr1_+_51191049 | 0.94 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr1_+_27023687 | 0.91 |
ENSDART00000189093
|
bnc2
|
basonuclin 2 |
| chr19_+_5543072 | 0.91 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr10_-_35257458 | 0.90 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr20_+_46513651 | 0.89 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr11_-_42554290 | 0.89 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr20_+_52442870 | 0.87 |
ENSDART00000163100
|
arhgap39
|
Rho GTPase activating protein 39 |
| chr8_+_49117518 | 0.86 |
ENSDART00000079631
|
rad21l1
|
RAD21 cohesin complex component like 1 |
| chr24_+_17269849 | 0.86 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr23_-_31913069 | 0.84 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr17_+_30545895 | 0.84 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr5_-_51619742 | 0.84 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr19_+_26423590 | 0.83 |
ENSDART00000136244
|
npr1a
|
natriuretic peptide receptor 1a |
| chr2_+_15048410 | 0.83 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr24_+_18948665 | 0.78 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr22_-_881725 | 0.77 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr11_+_39120306 | 0.77 |
ENSDART00000155746
ENSDART00000154158 |
cdc42
|
cell division cycle 42 |
| chr5_-_23117078 | 0.76 |
ENSDART00000051529
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr2_-_50225411 | 0.74 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr3_-_26805455 | 0.73 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr3_-_46811611 | 0.73 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr23_-_35790235 | 0.72 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr24_-_26399623 | 0.71 |
ENSDART00000112317
|
zgc:194621
|
zgc:194621 |
| chr17_-_21162821 | 0.70 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr19_-_7495006 | 0.70 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 13.8 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.6 | 4.8 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.3 | 5.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 1.1 | 4.4 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 1.1 | 10.8 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.8 | 2.4 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.8 | 2.4 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.6 | 2.4 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.5 | 4.9 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.5 | 2.7 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.5 | 8.8 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.5 | 1.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.4 | 1.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.4 | 10.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.4 | 6.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 1.4 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.3 | 1.7 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.3 | 7.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 2.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.3 | 10.7 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.3 | 1.3 | GO:0035994 | response to muscle stretch(GO:0035994) cardiac muscle fiber development(GO:0048739) |
| 0.3 | 0.9 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 1.1 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.2 | 11.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 4.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 5.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 1.9 | GO:0086002 | cardiac muscle cell action potential involved in contraction(GO:0086002) |
| 0.2 | 2.5 | GO:0042044 | fluid transport(GO:0042044) |
| 0.2 | 4.1 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.2 | 2.6 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.2 | 1.0 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.2 | 3.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 7.9 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 1.8 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.2 | 0.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 3.0 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.4 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.7 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.8 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 2.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 1.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 2.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 3.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.8 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 3.6 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 4.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 3.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 2.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.8 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 0.3 | GO:1903646 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 1.5 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 2.5 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 6.0 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.3 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 4.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.9 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.8 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 4.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 1.0 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 3.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 0.8 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 6.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.4 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 4.3 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 6.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.8 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 1.5 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
| 0.0 | 1.2 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 2.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.3 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.5 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 8.6 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 1.4 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 1.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 1.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 4.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 3.8 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 3.4 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 1.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.6 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 1.0 | GO:0031101 | fin regeneration(GO:0031101) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.1 | 10.8 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.8 | 4.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.6 | 2.3 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.5 | 5.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 4.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 1.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.3 | 5.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 8.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 3.0 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.9 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.9 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 2.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 21.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 4.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.3 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 23.9 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 6.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 4.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.5 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.6 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 2.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 2.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 7.5 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.1 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.7 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 6.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 6.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 4.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 18.7 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 1.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.6 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 13.8 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 1.4 | 7.2 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 1.1 | 3.2 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.8 | 2.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.6 | 10.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.6 | 2.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.6 | 1.7 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.4 | 4.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 8.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 1.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.3 | 10.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 4.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 4.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.3 | 4.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 1.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 4.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 1.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 2.5 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 1.0 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 2.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 6.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.2 | 1.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 8.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 6.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 6.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 3.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 4.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 2.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 5.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 1.4 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 0.8 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 5.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.7 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 1.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 2.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.7 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 3.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 3.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 1.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 7.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 6.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 2.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 1.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.9 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 1.7 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 6.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.9 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 7.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 6.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.6 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 6.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 8.0 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 2.2 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 4.7 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 9.5 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 2.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 4.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.7 | 4.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.5 | 8.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 6.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 6.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 1.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 9.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.0 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |