Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
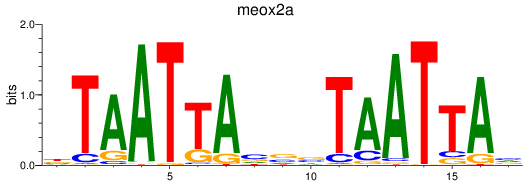
Results for meox2a
Z-value: 1.10
Transcription factors associated with meox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox2a
|
ENSDARG00000040911 | mesenchyme homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox2a | dr11_v1_chr15_-_34468599_34468599 | 0.05 | 6.5e-01 | Click! |
Activity profile of meox2a motif
Sorted Z-values of meox2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31267268 | 15.91 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr5_+_32222303 | 11.38 |
ENSDART00000051362
|
myhc4
|
myosin heavy chain 4 |
| chr6_+_55032439 | 10.22 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr23_-_27571667 | 8.31 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr23_+_26026383 | 7.86 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr15_+_45563656 | 6.68 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr2_-_30324610 | 6.56 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr15_+_45563491 | 6.12 |
ENSDART00000191169
|
cldn15lb
|
claudin 15-like b |
| chr5_+_2815021 | 5.72 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr6_-_58764672 | 5.67 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr19_+_9295244 | 5.51 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr11_-_2131280 | 5.04 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr12_-_26407092 | 4.89 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr8_-_13972626 | 4.85 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr5_-_26118855 | 4.59 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr8_+_39724138 | 4.53 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr16_+_12836143 | 4.51 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr5_-_63509581 | 4.40 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr20_-_40755614 | 4.12 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr18_+_7286788 | 3.91 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr8_-_10932206 | 3.86 |
ENSDART00000124313
|
nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr11_-_1509773 | 3.79 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr25_+_20077225 | 3.71 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr24_-_29963858 | 3.67 |
ENSDART00000183442
|
CR352310.1
|
|
| chr8_+_14886452 | 3.51 |
ENSDART00000146589
|
soat1
|
sterol O-acyltransferase 1 |
| chr10_+_26747755 | 3.46 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr6_-_32025225 | 3.38 |
ENSDART00000006417
|
pgm1
|
phosphoglucomutase 1 |
| chr5_+_23622177 | 3.30 |
ENSDART00000121504
|
cx27.5
|
connexin 27.5 |
| chr6_-_58757131 | 3.28 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr21_+_33187992 | 3.26 |
ENSDART00000162745
ENSDART00000188388 |
BX072577.1
|
|
| chr23_+_36106790 | 3.22 |
ENSDART00000128533
|
hoxc3a
|
homeobox C3a |
| chr2_+_24188502 | 3.19 |
ENSDART00000181955
ENSDART00000191706 ENSDART00000125909 |
map4l
|
microtubule associated protein 4 like |
| chr16_+_11585576 | 3.11 |
ENSDART00000172967
|
si:dkey-11o1.6
|
si:dkey-11o1.6 |
| chr1_+_12177195 | 3.08 |
ENSDART00000146842
ENSDART00000142081 |
stra6l
|
STRA6-like |
| chr14_-_3032016 | 3.04 |
ENSDART00000183461
ENSDART00000183035 |
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr14_-_30387894 | 2.97 |
ENSDART00000176136
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr4_+_5531583 | 2.95 |
ENSDART00000137500
ENSDART00000042080 |
si:dkey-14d8.6
|
si:dkey-14d8.6 |
| chr13_-_1408775 | 2.95 |
ENSDART00000049684
|
bag2
|
BCL2 associated athanogene 2 |
| chr2_+_50862527 | 2.87 |
ENSDART00000169800
ENSDART00000158847 ENSDART00000160900 |
adcyap1r1a
|
adenylate cyclase activating polypeptide 1a (pituitary) receptor type I |
| chr7_+_35075847 | 2.81 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr25_+_20089986 | 2.77 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr25_+_20091021 | 2.67 |
ENSDART00000187545
ENSDART00000053265 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr5_+_7279104 | 2.65 |
ENSDART00000190014
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr15_-_3252727 | 2.62 |
ENSDART00000131173
|
stoml3a
|
stomatin (EPB72)-like 3a |
| chr3_-_39152478 | 2.60 |
ENSDART00000154550
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr8_-_40555340 | 2.56 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr14_+_924876 | 2.49 |
ENSDART00000183908
|
myoz3a
|
myozenin 3a |
| chr23_-_11870962 | 2.47 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr22_+_15438872 | 2.47 |
ENSDART00000139800
|
gpc5b
|
glypican 5b |
| chr21_+_17880511 | 2.44 |
ENSDART00000080481
|
rxraa
|
retinoid X receptor, alpha a |
| chr24_-_7321928 | 2.41 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr3_+_2669813 | 2.37 |
ENSDART00000014205
|
CR388047.1
|
|
| chr14_+_22113331 | 2.36 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr6_-_11768198 | 2.35 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr7_-_5162292 | 2.31 |
ENSDART00000084218
|
zgc:195075
|
zgc:195075 |
| chr1_+_17593392 | 2.31 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr12_-_3753131 | 2.27 |
ENSDART00000129668
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr12_+_30586599 | 2.21 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr20_-_47051996 | 2.19 |
ENSDART00000153330
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr1_+_31110817 | 2.17 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr19_-_29887629 | 2.16 |
ENSDART00000066123
|
kpna6
|
karyopherin alpha 6 (importin alpha 7) |
| chr5_+_29820266 | 2.16 |
ENSDART00000146331
ENSDART00000098315 |
f11r.2
|
F11 receptor, tandem duplicate 2 |
| chr2_+_57801960 | 2.15 |
ENSDART00000147966
|
si:dkeyp-68b7.10
|
si:dkeyp-68b7.10 |
| chr19_+_5072918 | 2.15 |
ENSDART00000037126
|
eno2
|
enolase 2 |
| chr19_-_5805923 | 2.13 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr16_+_34479064 | 2.10 |
ENSDART00000159965
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr7_+_48705227 | 2.06 |
ENSDART00000174034
|
AL929208.1
|
|
| chr23_+_45538932 | 2.01 |
ENSDART00000135602
|
si:ch73-290k24.6
|
si:ch73-290k24.6 |
| chr23_-_44466257 | 2.00 |
ENSDART00000150126
|
si:ch1073-228j22.2
|
si:ch1073-228j22.2 |
| chr1_-_44701313 | 1.98 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr5_+_55984270 | 1.98 |
ENSDART00000047358
ENSDART00000138191 |
fkbp6
|
FK506 binding protein 6 |
| chr16_+_20161805 | 1.96 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr20_+_1960092 | 1.95 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr9_-_4606463 | 1.91 |
ENSDART00000179110
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr5_+_20147830 | 1.88 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr2_-_17392799 | 1.86 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr11_-_42750626 | 1.86 |
ENSDART00000130640
|
si:ch73-106k19.5
|
si:ch73-106k19.5 |
| chr7_+_14632157 | 1.86 |
ENSDART00000161264
|
ntrk3b
|
neurotrophic tyrosine kinase, receptor, type 3b |
| chr19_-_18313303 | 1.85 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr9_-_28616436 | 1.84 |
ENSDART00000136985
|
si:ch73-7i4.2
|
si:ch73-7i4.2 |
| chr5_+_20035284 | 1.83 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr23_+_16620801 | 1.82 |
ENSDART00000189859
ENSDART00000184578 |
snphb
|
syntaphilin b |
| chr19_-_28360033 | 1.82 |
ENSDART00000186994
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr1_-_12278056 | 1.82 |
ENSDART00000139336
ENSDART00000137463 |
cplx2l
|
complexin 2, like |
| chr5_-_68093169 | 1.81 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr24_+_14937205 | 1.80 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr15_+_5277761 | 1.78 |
ENSDART00000153954
|
si:ch1073-166e24.4
|
si:ch1073-166e24.4 |
| chr18_-_8857137 | 1.77 |
ENSDART00000126331
|
prrt4
|
proline-rich transmembrane protein 4 |
| chr6_-_33023745 | 1.76 |
ENSDART00000156211
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr21_+_29179887 | 1.75 |
ENSDART00000161941
|
si:ch211-57b15.1
|
si:ch211-57b15.1 |
| chr3_-_13946446 | 1.74 |
ENSDART00000171249
|
gcdhb
|
glutaryl-CoA dehydrogenase b |
| chr16_-_24642814 | 1.74 |
ENSDART00000153987
ENSDART00000154319 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr1_-_26063188 | 1.72 |
ENSDART00000168640
|
pdcd4a
|
programmed cell death 4a |
| chr6_-_55585423 | 1.71 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr20_-_6532462 | 1.70 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr25_-_4713461 | 1.70 |
ENSDART00000155302
|
drd4a
|
dopamine receptor D4a |
| chr5_+_25952340 | 1.69 |
ENSDART00000147188
|
trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr20_-_23171430 | 1.68 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr21_-_13123176 | 1.67 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr21_+_26612777 | 1.66 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr17_-_4252221 | 1.66 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr8_+_39795918 | 1.66 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr14_-_36320506 | 1.65 |
ENSDART00000074639
|
egf
|
epidermal growth factor |
| chr25_+_26895394 | 1.65 |
ENSDART00000155820
|
si:dkey-42p14.3
|
si:dkey-42p14.3 |
| chr9_+_46644633 | 1.62 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr19_+_31873308 | 1.62 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr9_+_2020667 | 1.58 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr1_+_25801648 | 1.58 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr17_-_43556415 | 1.57 |
ENSDART00000190102
ENSDART00000193156 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr10_+_26926654 | 1.56 |
ENSDART00000078980
ENSDART00000100289 |
rab1bb
|
RAB1B, member RAS oncogene family b |
| chr2_+_10709710 | 1.55 |
ENSDART00000154712
|
evi5a
|
ecotropic viral integration site 5a |
| chr12_+_18681477 | 1.55 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr25_+_2361721 | 1.54 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr2_-_17393216 | 1.54 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr9_-_31596016 | 1.54 |
ENSDART00000142289
|
nalcn
|
sodium leak channel, non-selective |
| chr18_+_5137241 | 1.54 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr9_-_48407408 | 1.53 |
ENSDART00000058248
|
zgc:172182
|
zgc:172182 |
| chr5_+_28497956 | 1.53 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr2_+_50608099 | 1.52 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr25_-_10565006 | 1.52 |
ENSDART00000130608
ENSDART00000190212 |
galn
|
galanin/GMAP prepropeptide |
| chr25_-_8030113 | 1.50 |
ENSDART00000104674
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr15_-_7598294 | 1.49 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr15_-_44077937 | 1.48 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr18_+_11506561 | 1.47 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr22_+_17205608 | 1.46 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr11_+_22374419 | 1.45 |
ENSDART00000174683
ENSDART00000170521 ENSDART00000193980 |
ppfia4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr13_-_30713236 | 1.45 |
ENSDART00000112372
ENSDART00000142221 |
tmem72
|
transmembrane protein 72 |
| chr17_+_33495194 | 1.44 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr13_+_17702522 | 1.43 |
ENSDART00000057913
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr13_-_37608441 | 1.43 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr3_+_13624815 | 1.42 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr2_-_2957970 | 1.42 |
ENSDART00000162505
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr6_+_2195625 | 1.42 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr16_-_24605969 | 1.41 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr4_-_77506362 | 1.37 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr15_+_44366556 | 1.36 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr9_-_51323545 | 1.35 |
ENSDART00000139316
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr1_-_45920632 | 1.34 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr14_+_3287740 | 1.33 |
ENSDART00000186290
|
cdx1a
|
caudal type homeobox 1a |
| chr16_+_14588141 | 1.33 |
ENSDART00000140469
ENSDART00000059984 ENSDART00000167411 ENSDART00000133566 |
deptor
|
DEP domain containing MTOR-interacting protein |
| chr20_+_26966725 | 1.33 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr19_-_8768564 | 1.31 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr7_+_47243564 | 1.31 |
ENSDART00000098942
ENSDART00000162237 |
znf507
|
zinc finger protein 507 |
| chr3_-_19091024 | 1.30 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr24_+_31655939 | 1.30 |
ENSDART00000187337
|
CABZ01029422.1
|
|
| chr21_+_15824182 | 1.30 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr14_+_25465346 | 1.30 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr1_-_10647484 | 1.29 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr18_+_9615147 | 1.28 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr14_-_26094172 | 1.28 |
ENSDART00000150131
|
mtnr1al
|
melatonin receptor type 1A like |
| chr13_-_23264724 | 1.28 |
ENSDART00000051886
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr3_+_22036391 | 1.26 |
ENSDART00000147721
|
cdc27
|
cell division cycle 27 |
| chr20_+_19816465 | 1.26 |
ENSDART00000079613
|
chrna2b
|
cholinergic receptor, nicotinic, alpha 2b (neuronal) |
| chr10_+_21576909 | 1.25 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr23_+_4646194 | 1.25 |
ENSDART00000092344
|
LO017700.1
|
|
| chr2_+_34572690 | 1.24 |
ENSDART00000077216
|
astn1
|
astrotactin 1 |
| chr17_+_30751462 | 1.24 |
ENSDART00000154184
|
ldah
|
lipid droplet associated hydrolase |
| chr6_+_49412754 | 1.24 |
ENSDART00000027398
|
kcna2a
|
potassium voltage-gated channel, shaker-related subfamily, member 2a |
| chr7_+_65261576 | 1.23 |
ENSDART00000169566
|
bco1
|
beta-carotene oxygenase 1 |
| chr3_-_40054615 | 1.23 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr19_-_5865766 | 1.23 |
ENSDART00000191007
|
LO018585.1
|
|
| chr8_+_3434146 | 1.23 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr4_-_2168805 | 1.22 |
ENSDART00000150039
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr24_+_33802528 | 1.22 |
ENSDART00000136040
ENSDART00000147499 ENSDART00000182322 |
atg9b
|
autophagy related 9B |
| chr16_+_23598908 | 1.21 |
ENSDART00000131627
|
kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr7_-_27685365 | 1.20 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr9_+_49712868 | 1.20 |
ENSDART00000192969
ENSDART00000183310 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr9_-_34882516 | 1.19 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr7_+_35245607 | 1.19 |
ENSDART00000193422
ENSDART00000173888 |
amfrb
|
autocrine motility factor receptor b |
| chr8_+_49778486 | 1.19 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr11_-_7078392 | 1.18 |
ENSDART00000112156
ENSDART00000188556 |
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr21_-_26071773 | 1.17 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr17_+_10090638 | 1.17 |
ENSDART00000169522
ENSDART00000160156 |
sec23a
|
Sec23 homolog A, COPII coat complex component |
| chr4_-_48727718 | 1.17 |
ENSDART00000121499
|
znf1023
|
zinc finger protein 1023 |
| chr5_+_69981453 | 1.16 |
ENSDART00000143860
|
si:ch211-154e10.1
|
si:ch211-154e10.1 |
| chr3_+_431208 | 1.16 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr2_+_44972720 | 1.16 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr2_+_56937548 | 1.15 |
ENSDART00000189308
|
CABZ01117752.1
|
|
| chr3_+_52528606 | 1.14 |
ENSDART00000111510
|
ptger1c
|
prostaglandin E receptor 1c (subtype EP1) |
| chr2_-_14390627 | 1.14 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr3_-_32169754 | 1.13 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr3_-_24458281 | 1.12 |
ENSDART00000153993
|
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr9_-_54248182 | 1.12 |
ENSDART00000129540
|
larsa
|
leucyl-tRNA synthetase a |
| chr10_-_2524917 | 1.11 |
ENSDART00000188642
|
CU856539.1
|
|
| chr23_+_14696043 | 1.10 |
ENSDART00000132037
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr22_-_12746539 | 1.10 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr23_-_306796 | 1.09 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr6_-_31325400 | 1.09 |
ENSDART00000188869
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr7_-_34294613 | 1.08 |
ENSDART00000191632
|
CR388195.1
|
|
| chr21_-_18648861 | 1.08 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr19_+_14059349 | 1.08 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr25_-_8030425 | 1.08 |
ENSDART00000014964
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr9_+_22656976 | 1.07 |
ENSDART00000136249
ENSDART00000139270 |
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr16_-_27174373 | 1.07 |
ENSDART00000166681
|
frrs1l
|
ferric-chelate reductase 1-like |
| chr14_+_36628131 | 1.07 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr17_-_6618574 | 1.05 |
ENSDART00000184486
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr10_+_5203532 | 1.05 |
ENSDART00000165018
|
cdc42se2
|
CDC42 small effector 2 |
| chr1_+_47178529 | 1.05 |
ENSDART00000158432
ENSDART00000074450 ENSDART00000137448 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr24_+_9178064 | 1.04 |
ENSDART00000142971
|
dlgap1b
|
discs, large (Drosophila) homolog-associated protein 1b |
| chr10_-_8053385 | 1.03 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 1.4 | 12.5 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 1.0 | 5.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.9 | 4.5 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.8 | 8.3 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.6 | 3.1 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.5 | 1.6 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.5 | 1.6 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.5 | 2.0 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.5 | 26.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.5 | 3.0 | GO:1902837 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.5 | 7.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.5 | 1.4 | GO:0098581 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.4 | 11.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.4 | 1.6 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.4 | 1.2 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.4 | 2.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.4 | 1.1 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.4 | 1.8 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 1.8 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.4 | 1.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 2.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 0.7 | GO:0046635 | positive regulation of alpha-beta T cell activation(GO:0046635) |
| 0.3 | 1.7 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.3 | 4.5 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.3 | 0.9 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.3 | 1.7 | GO:0016048 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.3 | 3.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 1.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.3 | 2.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 2.0 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 1.7 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 2.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 0.7 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.2 | 1.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 1.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.2 | 0.6 | GO:0007567 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.2 | 0.8 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 1.0 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.2 | 1.0 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.2 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 2.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.7 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.2 | 0.8 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.2 | 1.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 2.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.2 | 0.5 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.1 | 0.4 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 1.3 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 0.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.7 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 3.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.4 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 1.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 1.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 1.7 | GO:0051967 | spinal cord motor neuron cell fate specification(GO:0021520) negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 1.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.9 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.1 | 0.7 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 3.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 1.2 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 1.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.8 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.9 | GO:0034672 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.1 | 1.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.6 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 2.2 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 0.5 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 1.3 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.3 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.4 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.6 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 4.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 2.9 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 5.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.6 | GO:0031646 | positive regulation of myelination(GO:0031643) positive regulation of neurological system process(GO:0031646) |
| 0.1 | 2.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.4 | GO:1900044 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 1.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.8 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 1.3 | GO:0099565 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 1.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.4 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 1.0 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 1.3 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 2.0 | GO:0019915 | lipid storage(GO:0019915) |
| 0.1 | 0.1 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.8 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 2.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.7 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.1 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of nucleotide metabolic process(GO:0045980) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) negative regulation of purine nucleotide metabolic process(GO:1900543) |
| 0.0 | 3.5 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.5 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 1.8 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 3.5 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 2.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.8 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.6 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 1.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.4 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 2.6 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 2.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.7 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.6 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 1.6 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.3 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.5 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 1.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.4 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 1.1 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 1.0 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.6 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.6 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 1.2 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.5 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.6 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 1.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 2.1 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 1.0 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.4 | GO:0043506 | activation of JUN kinase activity(GO:0007257) regulation of JUN kinase activity(GO:0043506) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.9 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.6 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 1.0 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.2 | GO:0001754 | eye photoreceptor cell differentiation(GO:0001754) |
| 0.0 | 2.6 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 1.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.0 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 1.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.0 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 2.0 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.6 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.3 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 1.6 | GO:0042391 | regulation of membrane potential(GO:0042391) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.9 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 1.3 | 6.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.8 | 8.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.6 | 4.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.6 | 26.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 1.6 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.3 | 2.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 0.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 1.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 0.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 1.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 3.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 1.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 1.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.0 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 13.5 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.4 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 1.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 11.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 3.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 4.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.1 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 19.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.6 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.5 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 1.9 | 5.7 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.8 | 8.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.7 | 7.4 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.6 | 7.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.5 | 3.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 3.4 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.5 | 2.9 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.5 | 1.4 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.4 | 1.7 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.4 | 1.3 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.4 | 3.0 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 2.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.4 | 1.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.4 | 3.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.4 | 1.5 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.4 | 1.8 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 2.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 3.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 2.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 1.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 3.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.3 | 2.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 2.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 1.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 2.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 5.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 0.6 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.2 | 0.8 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 1.0 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.2 | 1.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.2 | 0.9 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 0.7 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.2 | 4.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 7.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.9 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 3.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 1.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 2.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 3.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 3.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 0.3 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.6 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.7 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.1 | 0.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 3.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.4 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.1 | 0.7 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.1 | 1.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.6 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.1 | 0.6 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 1.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.5 | GO:0038187 | pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.7 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 7.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 2.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.1 | 0.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 0.9 | GO:0019870 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 2.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 5.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 11.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.6 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.5 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.5 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.8 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 2.6 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.9 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 2.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.2 | 1.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 3.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.5 | 11.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 2.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 4.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 3.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 1.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 4.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 0.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.7 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.4 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 3.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.2 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |