Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
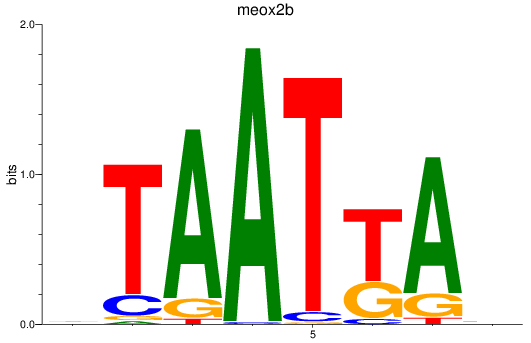
Results for meox2b
Z-value: 0.92
Transcription factors associated with meox2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox2b
|
ENSDARG00000061818 | mesenchyme homeobox 2b |
|
meox2b
|
ENSDARG00000116206 | mesenchyme homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox2b | dr11_v1_chr19_+_31183495_31183495 | -0.07 | 5.1e-01 | Click! |
Activity profile of meox2b motif
Sorted Z-values of meox2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_21867307 | 18.50 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr20_-_40755614 | 14.23 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr21_+_27416284 | 13.63 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr12_+_31713239 | 12.31 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr16_+_23913943 | 11.37 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr12_-_22524388 | 9.45 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr6_-_27123327 | 8.84 |
ENSDART00000073881
|
agxta
|
alanine-glyoxylate aminotransferase a |
| chr15_-_21877726 | 8.62 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr5_-_25723079 | 8.47 |
ENSDART00000014013
|
gda
|
guanine deaminase |
| chr24_-_38110779 | 8.40 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr13_-_20381485 | 7.89 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr22_-_24738188 | 7.89 |
ENSDART00000050238
|
vtg1
|
vitellogenin 1 |
| chr6_+_41191482 | 7.71 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr12_-_35830625 | 7.51 |
ENSDART00000180028
|
CU459056.1
|
|
| chr8_-_10932206 | 7.44 |
ENSDART00000124313
|
nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr15_+_45563656 | 7.14 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr16_-_54455573 | 7.05 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr22_+_12770877 | 7.02 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr16_-_2414063 | 7.02 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr16_-_45178430 | 6.64 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr18_+_15644559 | 6.44 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr4_-_2380173 | 6.37 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr15_+_36457888 | 6.30 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr16_+_23431189 | 5.81 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr25_-_13188678 | 5.77 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr22_-_15578402 | 5.74 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr7_+_21841037 | 5.71 |
ENSDART00000077503
|
tm4sf5
|
transmembrane 4 L six family member 5 |
| chr3_+_17537352 | 5.71 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr25_+_10410620 | 5.62 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr5_-_42904329 | 5.49 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr22_+_1170294 | 5.45 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr11_-_37997419 | 5.22 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr10_+_25726694 | 5.19 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr1_+_53374454 | 5.14 |
ENSDART00000038807
|
ucp1
|
uncoupling protein 1 |
| chr13_+_23988442 | 5.11 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr14_-_15171435 | 5.09 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr13_+_33688474 | 5.04 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr7_+_54642005 | 5.01 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr1_-_45049603 | 4.94 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr8_+_14886452 | 4.80 |
ENSDART00000146589
|
soat1
|
sterol O-acyltransferase 1 |
| chr16_+_29509133 | 4.77 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr16_-_17197546 | 4.76 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr8_+_6576940 | 4.73 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr3_+_13624815 | 4.72 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr14_-_21064199 | 4.70 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr15_+_46357080 | 4.70 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr2_-_5475910 | 4.57 |
ENSDART00000100954
ENSDART00000172143 ENSDART00000132496 |
proca
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr13_-_8692860 | 4.54 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr25_+_3327071 | 4.52 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr19_-_11237125 | 4.48 |
ENSDART00000163921
|
ssr2
|
signal sequence receptor, beta |
| chr20_-_49704915 | 4.44 |
ENSDART00000189232
|
COX7A2 (1 of many)
|
cytochrome c oxidase subunit 7A2 |
| chr23_+_27703749 | 4.26 |
ENSDART00000027224
|
lmbr1l
|
limb development membrane protein 1-like |
| chr22_-_24818066 | 4.25 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr5_+_27897504 | 4.23 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr7_+_73447724 | 4.21 |
ENSDART00000040786
|
cbln6
|
cerebellin 6 |
| chr16_+_23984179 | 4.15 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr3_+_18398876 | 4.14 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr21_-_5205617 | 4.12 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr21_+_25777425 | 4.12 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr13_-_33170733 | 4.08 |
ENSDART00000057382
|
fbln5
|
fibulin 5 |
| chr21_-_19919020 | 4.02 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr13_-_8692432 | 3.98 |
ENSDART00000058106
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr1_-_23308225 | 3.97 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr7_+_69019851 | 3.96 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr3_+_1015867 | 3.95 |
ENSDART00000109912
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr24_-_11076400 | 3.95 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr19_-_11238620 | 3.94 |
ENSDART00000148697
ENSDART00000149169 |
ssr2
|
signal sequence receptor, beta |
| chr17_-_30635298 | 3.92 |
ENSDART00000155478
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr10_-_3295197 | 3.89 |
ENSDART00000109131
|
slc25a1b
|
slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b |
| chr4_+_391297 | 3.85 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr13_+_28747934 | 3.85 |
ENSDART00000160176
ENSDART00000101633 ENSDART00000136318 |
prom2
|
prominin 2 |
| chr14_+_35428152 | 3.84 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr10_-_24391716 | 3.82 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr20_+_2281933 | 3.78 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr13_+_25720969 | 3.74 |
ENSDART00000046050
|
pcbd1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr11_+_1602916 | 3.73 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr17_+_20174812 | 3.70 |
ENSDART00000186628
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr1_-_23293261 | 3.67 |
ENSDART00000122648
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr19_-_42556086 | 3.65 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr7_-_12968689 | 3.65 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr16_+_42471455 | 3.64 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr15_+_46356879 | 3.64 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr14_+_32942063 | 3.62 |
ENSDART00000187705
|
lnx2b
|
ligand of numb-protein X 2b |
| chr16_-_25606235 | 3.61 |
ENSDART00000192741
|
zgc:110410
|
zgc:110410 |
| chr10_-_8046764 | 3.60 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr20_+_36629173 | 3.57 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr2_-_26596794 | 3.53 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr16_+_17714664 | 3.52 |
ENSDART00000149042
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr20_-_23426339 | 3.50 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr18_-_19456269 | 3.50 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr19_-_25081711 | 3.49 |
ENSDART00000058513
|
xkr8.3
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 3 |
| chr1_+_52392511 | 3.49 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr9_-_712308 | 3.46 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr16_+_33902006 | 3.42 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr1_-_58562129 | 3.42 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr2_+_22851832 | 3.41 |
ENSDART00000145944
|
amotl2b
|
angiomotin like 2b |
| chr6_+_8630355 | 3.37 |
ENSDART00000161749
ENSDART00000193976 |
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr10_-_21955231 | 3.36 |
ENSDART00000183695
|
FO744833.2
|
|
| chr2_-_10703621 | 3.35 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr4_+_14900042 | 3.34 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr11_-_45138857 | 3.33 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr6_-_43283122 | 3.33 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr7_-_38792543 | 3.33 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr16_-_29387215 | 3.32 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr8_-_46525092 | 3.26 |
ENSDART00000030482
|
sult1st2
|
sulfotransferase family 1, cytosolic sulfotransferase 2 |
| chr11_-_28050559 | 3.25 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr21_+_19062124 | 3.24 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr4_-_13502549 | 3.24 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr1_-_9195629 | 3.22 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr10_-_8053753 | 3.20 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr18_+_14564085 | 3.19 |
ENSDART00000009363
ENSDART00000141813 ENSDART00000136120 |
si:dkey-246g23.4
|
si:dkey-246g23.4 |
| chr21_+_1647990 | 3.17 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr12_+_20352400 | 3.17 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr14_+_41345175 | 3.17 |
ENSDART00000086104
|
nox1
|
NADPH oxidase 1 |
| chr10_+_8690936 | 3.16 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr7_+_26545502 | 3.14 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr4_-_9891874 | 3.13 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr21_-_19314618 | 3.12 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr5_-_33287691 | 3.12 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr5_-_69620722 | 3.11 |
ENSDART00000097248
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr8_-_27656765 | 3.11 |
ENSDART00000078491
|
mov10b.2
|
Moloney leukemia virus 10b, tandem duplicate 2 |
| chr18_+_22109379 | 3.11 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr21_+_43702016 | 3.07 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr13_+_7442023 | 3.04 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr16_+_16969060 | 3.03 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr10_-_8060573 | 3.01 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr6_-_1591002 | 3.00 |
ENSDART00000087039
|
zgc:123305
|
zgc:123305 |
| chr1_-_43905252 | 2.99 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr23_-_306796 | 2.98 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr14_-_30387894 | 2.97 |
ENSDART00000176136
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr23_+_39695827 | 2.97 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr1_-_58561963 | 2.97 |
ENSDART00000165040
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr7_+_20031202 | 2.95 |
ENSDART00000052904
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr17_-_21842135 | 2.95 |
ENSDART00000154927
|
cuzd1.2
|
CUB and zona pellucida-like domains 1, tandem duplicate 2 |
| chr8_+_15254564 | 2.94 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr5_+_19343880 | 2.93 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr7_-_54679595 | 2.91 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr20_+_15015557 | 2.89 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr21_-_5881344 | 2.89 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr5_+_26213874 | 2.88 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr3_-_33427803 | 2.86 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr11_+_40812590 | 2.86 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr16_-_51271962 | 2.86 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr18_+_7073130 | 2.85 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr16_-_9675982 | 2.83 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr17_+_8799451 | 2.81 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr12_+_20587179 | 2.79 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr19_-_41213718 | 2.79 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr19_+_770300 | 2.79 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr20_+_41021054 | 2.79 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr6_-_55399214 | 2.77 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr22_+_20427170 | 2.74 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr5_+_32882688 | 2.72 |
ENSDART00000008807
ENSDART00000185317 |
rpl12
|
ribosomal protein L12 |
| chr25_-_13703826 | 2.70 |
ENSDART00000163398
|
pla2g15
|
phospholipase A2, group XV |
| chr10_-_21362320 | 2.70 |
ENSDART00000189789
|
avd
|
avidin |
| chr11_+_16216909 | 2.68 |
ENSDART00000081035
ENSDART00000147190 |
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr10_-_8079737 | 2.68 |
ENSDART00000059014
ENSDART00000179549 |
zgc:173443
si:ch211-251f6.6
|
zgc:173443 si:ch211-251f6.6 |
| chr25_+_3326885 | 2.67 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr10_-_21362071 | 2.67 |
ENSDART00000125167
|
avd
|
avidin |
| chr24_-_25166720 | 2.66 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr13_+_29238850 | 2.65 |
ENSDART00000026000
|
myofl
|
myoferlin like |
| chr6_+_6828167 | 2.64 |
ENSDART00000181284
|
si:ch211-85n16.4
|
si:ch211-85n16.4 |
| chr20_-_6532462 | 2.63 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr22_-_16154771 | 2.62 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr24_-_41267184 | 2.61 |
ENSDART00000063504
|
xylb
|
xylulokinase homolog (H. influenzae) |
| chr23_+_2906031 | 2.59 |
ENSDART00000109304
|
c23h20orf24
|
c23h20orf24 homolog (H. sapiens) |
| chr10_+_6884627 | 2.59 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr9_+_50316921 | 2.58 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr15_+_26603395 | 2.57 |
ENSDART00000188667
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr25_+_29474982 | 2.57 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr24_+_19415124 | 2.56 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr22_-_10586191 | 2.56 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr18_-_33979422 | 2.55 |
ENSDART00000136535
ENSDART00000167698 |
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr14_-_32631013 | 2.54 |
ENSDART00000176815
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr16_-_41762983 | 2.54 |
ENSDART00000192936
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr11_+_44135351 | 2.53 |
ENSDART00000182914
|
FO704721.1
|
|
| chr10_+_29850330 | 2.52 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr25_-_21031007 | 2.51 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr18_+_20560616 | 2.50 |
ENSDART00000136710
ENSDART00000151974 ENSDART00000121699 ENSDART00000040074 |
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr1_+_51615672 | 2.49 |
ENSDART00000165117
|
zgc:165656
|
zgc:165656 |
| chr3_-_16719244 | 2.48 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr13_-_31017960 | 2.48 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr24_-_37640705 | 2.47 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr10_+_11261576 | 2.46 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr10_-_44560165 | 2.44 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr12_-_3940768 | 2.43 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr3_-_61494840 | 2.41 |
ENSDART00000101957
|
baiap2l1b
|
BAI1-associated protein 2-like 1b |
| chr13_-_40238813 | 2.41 |
ENSDART00000044963
|
loxl4
|
lysyl oxidase-like 4 |
| chr5_+_23242370 | 2.41 |
ENSDART00000051532
|
agtr2
|
angiotensin II receptor, type 2 |
| chr1_-_43727012 | 2.41 |
ENSDART00000181064
|
bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr24_+_21621654 | 2.41 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr1_-_999556 | 2.41 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr9_-_2892045 | 2.40 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr18_+_3037998 | 2.40 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr24_+_16985181 | 2.40 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr7_-_64589920 | 2.39 |
ENSDART00000172619
ENSDART00000184113 |
CR387919.1
|
|
| chr12_+_48803098 | 2.39 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr13_-_25720876 | 2.38 |
ENSDART00000142404
ENSDART00000146487 |
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr15_+_31344472 | 2.38 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr9_-_2892250 | 2.38 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 2.3 | 7.0 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 2.0 | 12.1 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 1.7 | 8.6 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 1.7 | 6.6 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 1.6 | 4.7 | GO:0098581 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 1.5 | 8.8 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 1.3 | 4.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 1.3 | 4.0 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 1.0 | 3.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 1.0 | 3.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 1.0 | 3.9 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.9 | 4.7 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.9 | 7.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.9 | 2.6 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.9 | 3.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.8 | 4.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.8 | 2.3 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.8 | 2.3 | GO:0044038 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.7 | 2.9 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.7 | 2.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.7 | 3.4 | GO:0035912 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.7 | 2.0 | GO:0030238 | male sex determination(GO:0030238) |
| 0.7 | 2.0 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.6 | 3.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.6 | 1.9 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.6 | 3.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.6 | 2.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.6 | 2.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.6 | 2.3 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.6 | 2.9 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.6 | 2.3 | GO:0060471 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.6 | 2.8 | GO:0045056 | transcytosis(GO:0045056) |
| 0.5 | 1.6 | GO:0036316 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.5 | 2.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.5 | 3.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.5 | 2.0 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.5 | 3.0 | GO:1902765 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.5 | 1.5 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.5 | 2.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.5 | 2.4 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.5 | 2.4 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.5 | 5.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.5 | 2.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.5 | 5.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.5 | 2.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 4.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.4 | 1.3 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.4 | 3.0 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.4 | 2.5 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.4 | 2.4 | GO:0097065 | anterior head development(GO:0097065) |
| 0.4 | 2.0 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.4 | 6.4 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.4 | 2.4 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.4 | 3.2 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.4 | 1.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.4 | 2.6 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.4 | 3.7 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.4 | 1.9 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.4 | 1.1 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.4 | 7.6 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.4 | 13.4 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.3 | 2.8 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.3 | 2.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 1.4 | GO:0014005 | microglia development(GO:0014005) |
| 0.3 | 1.0 | GO:1903726 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.3 | 1.6 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.3 | 2.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 0.9 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.3 | 3.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 5.1 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.3 | 3.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.3 | 2.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 1.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 8.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.3 | 1.4 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.3 | 1.3 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.3 | 3.8 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.3 | 1.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 1.2 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 4.8 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.2 | 1.9 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.2 | 1.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 1.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 0.9 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.2 | 0.9 | GO:0006562 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) proline catabolic process(GO:0006562) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 3.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 1.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 3.7 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.2 | 2.3 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.2 | 0.9 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 2.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 0.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 0.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.2 | 1.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 0.9 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 5.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 1.3 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.2 | 7.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.2 | 9.2 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 3.1 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 2.7 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 16.2 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.2 | 1.4 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 6.1 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.2 | 0.8 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 1.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 0.6 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.2 | 2.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 1.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 0.6 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.2 | 5.0 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.2 | 2.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 0.5 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) RNA 5'-end processing(GO:0000966) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 2.6 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.2 | 1.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 2.9 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.2 | 0.7 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 3.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 1.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 0.5 | GO:0010656 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.2 | 0.9 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.2 | 0.5 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.2 | 2.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 0.8 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.2 | 1.1 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.2 | 2.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.2 | 2.6 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.7 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.2 | 3.8 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.2 | 3.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 0.9 | GO:0032042 | mitochondrial DNA replication(GO:0006264) mitochondrial DNA metabolic process(GO:0032042) |
| 0.2 | 0.3 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 4.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 2.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 3.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 1.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.9 | GO:0097101 | blood vessel endothelial cell fate commitment(GO:0060846) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 4.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 7.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 2.4 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 1.7 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.8 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 13.5 | GO:0072376 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.1 | 1.5 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 2.9 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.2 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 2.3 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 1.3 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 3.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.8 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.4 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 0.4 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 1.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.5 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.9 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 3.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 1.6 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 0.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 1.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.6 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 2.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.7 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 1.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 5.9 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 1.0 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 1.0 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.6 | GO:0003203 | endocardial cushion morphogenesis(GO:0003203) endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.1 | 0.9 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.9 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.1 | 1.9 | GO:1904018 | positive regulation of angiogenesis(GO:0045766) positive regulation of vasculature development(GO:1904018) |
| 0.1 | 2.3 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.1 | 1.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.4 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.1 | 1.6 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 0.5 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.9 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 1.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.6 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 1.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.7 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 2.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 6.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 2.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.8 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 1.8 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 1.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 2.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.1 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 2.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.0 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 5.7 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.1 | 0.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 2.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.7 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 6.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 2.3 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 2.7 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 34.4 | GO:0043043 | translation(GO:0006412) peptide biosynthetic process(GO:0043043) |
| 0.1 | 1.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.9 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 4.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 13.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.1 | 0.6 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.2 | GO:0046823 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.1 | 1.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.4 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 2.0 | GO:0043434 | response to peptide hormone(GO:0043434) |
| 0.1 | 1.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 1.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.2 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 1.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.8 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 0.8 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.0 | GO:0060173 | limb development(GO:0060173) |
| 0.1 | 2.6 | GO:0010952 | positive regulation of endopeptidase activity(GO:0010950) positive regulation of peptidase activity(GO:0010952) |
| 0.1 | 0.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 2.6 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.6 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 1.3 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.9 | GO:0031529 | ruffle organization(GO:0031529) ruffle assembly(GO:0097178) |
| 0.0 | 0.2 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 1.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.0 | 0.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.6 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 1.7 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.7 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 1.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.9 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 2.0 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.9 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 1.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0046386 | deoxyribonucleotide catabolic process(GO:0009264) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.0 | 0.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.5 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.0 | 2.4 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 1.6 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 4.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.6 | GO:1901661 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 2.0 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 2.3 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.2 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.2 | GO:0072178 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.0 | 1.5 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.0 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 0.7 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 2.0 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.0 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.6 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 12.2 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 1.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.9 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 1.2 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 3.8 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.8 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 1.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.7 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 1.2 | GO:0036293 | response to decreased oxygen levels(GO:0036293) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.3 | GO:0014902 | myotube differentiation(GO:0014902) |
| 0.0 | 0.2 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 1.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.1 | GO:0097067 | response to thyroid hormone(GO:0097066) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.3 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.8 | GO:0051961 | negative regulation of neurogenesis(GO:0050768) negative regulation of nervous system development(GO:0051961) |
| 0.0 | 0.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 3.5 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.6 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 1.5 | GO:0045165 | cell fate commitment(GO:0045165) |
| 0.0 | 0.6 | GO:0071774 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 2.5 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.7 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.7 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.2 | GO:0001946 | lymphangiogenesis(GO:0001946) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.0 | 12.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.8 | 3.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.7 | 2.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.6 | 2.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.5 | 2.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.5 | 2.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.5 | 1.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.5 | 32.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.5 | 2.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.5 | 3.8 | GO:0071914 | prominosome(GO:0071914) |
| 0.5 | 2.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.5 | 1.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.4 | 1.3 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.4 | 1.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 2.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 3.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 1.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 3.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 2.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 9.4 | GO:0043186 | P granule(GO:0043186) |
| 0.3 | 4.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 3.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.2 | 8.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 26.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.2 | 0.8 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.2 | 2.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 0.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 7.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 4.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 2.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 5.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 0.9 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 5.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 2.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 6.5 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 0.9 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 12.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 12.4 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 1.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 2.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 3.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 7.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 9.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 3.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.9 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 1.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.7 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.5 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 4.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 3.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 5.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 3.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.5 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 9.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 18.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 27.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.5 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 1.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 3.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) COPI-coated vesicle(GO:0030137) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 8.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.7 | 12.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 1.7 | 8.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.7 | 6.6 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 1.6 | 4.7 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 1.4 | 5.5 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 1.3 | 5.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.3 | 3.8 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 1.2 | 4.8 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.2 | 3.6 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 1.1 | 5.4 | GO:0009374 | biotin binding(GO:0009374) |
| 1.1 | 6.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 1.0 | 3.1 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 1.0 | 5.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.9 | 5.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.9 | 3.7 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.9 | 6.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.8 | 5.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.7 | 2.9 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.7 | 2.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.7 | 3.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.7 | 2.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.6 | 1.9 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.6 | 2.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.6 | 2.4 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.6 | 2.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.5 | 2.1 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.5 | 4.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.5 | 3.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.5 | 4.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.5 | 2.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 2.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.5 | 1.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.4 | 2.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.4 | 2.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.4 | 13.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.4 | 1.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.4 | 1.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.4 | 2.0 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.4 | 1.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 2.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.4 | 1.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 1.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.4 | 1.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 10.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.4 | 1.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.4 | 1.4 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.4 | 3.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 2.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 4.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 2.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 1.3 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.3 | 0.9 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.3 | 3.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.3 | 1.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 1.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 7.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.3 | 12.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.3 | 2.4 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.3 | 3.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 1.1 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.3 | 1.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.3 | 2.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 1.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 5.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 2.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 1.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 0.7 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 2.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.2 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.2 | 0.9 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.2 | 10.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.2 | 2.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.2 | 3.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 40.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 1.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 3.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 1.8 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 5.3 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 0.6 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 8.5 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.2 | 2.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 0.6 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.2 | 2.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 2.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 2.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 1.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 40.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 5.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 1.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.2 | 2.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 2.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 1.2 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 3.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 7.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 2.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 2.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.2 | 2.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 3.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 3.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.7 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 2.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.8 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 2.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.5 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 7.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 3.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.3 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.8 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.8 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.1 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 1.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 7.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.9 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 7.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.8 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 2.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 2.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 0.5 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 4.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 1.6 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.1 | 1.0 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 4.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 8.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.9 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.9 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 6.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.1 | 1.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 1.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.0 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.1 | 2.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 1.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.5 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 1.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 1.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.5 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 3.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.5 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 2.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 7.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 3.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 7.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 1.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.7 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 4.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.8 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 2.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0043047 | double-stranded telomeric DNA binding(GO:0003691) single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 1.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.2 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 4.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 1.0 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.4 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.0 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.7 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 13.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 1.0 | 4.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.4 | 4.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 9.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.3 | 8.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 6.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 4.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 1.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 3.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 2.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 8.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 1.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 27.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 5.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 2.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 0.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 2.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 6.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 6.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 13.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.7 | 8.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.8 | 8.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.7 | 6.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.6 | 11.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 4.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 45.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.4 | 2.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 8.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 3.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 7.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 8.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 8.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.2 | 3.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 2.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 3.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 2.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 5.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 1.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 2.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 1.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 8.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 2.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 1.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 13.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 3.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 3.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 3.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 7.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 3.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.2 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 0.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 4.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 13.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 4.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 0.9 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.7 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 2.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.9 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE EXCISION REPAIR | Genes involved in Nucleotide Excision Repair |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |