Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
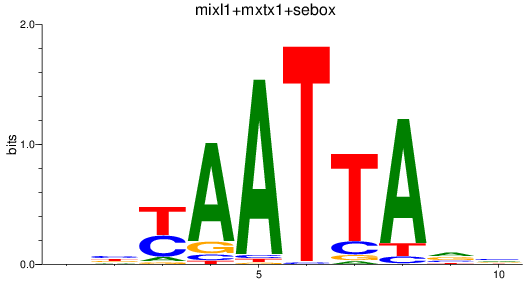
Results for mixl1+mxtx1+sebox
Z-value: 0.55
Transcription factors associated with mixl1+mxtx1+sebox
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sebox
|
ENSDARG00000042526 | SEBOX homeobox |
|
mixl1
|
ENSDARG00000069252 | Mix paired-like homeobox |
|
mxtx1
|
ENSDARG00000069382 | mix-type homeobox gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mxtx1 | dr11_v1_chr13_-_21660203_21660203 | -0.17 | 1.1e-01 | Click! |
| mixl1 | dr11_v1_chr20_-_43723860_43723860 | 0.05 | 6.3e-01 | Click! |
| sebox | dr11_v1_chr5_+_67371650_67371650 | -0.03 | 7.4e-01 | Click! |
Activity profile of mixl1+mxtx1+sebox motif
Sorted Z-values of mixl1+mxtx1+sebox motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_41494831 | 6.31 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr6_-_54826061 | 4.21 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr25_+_31276842 | 3.75 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr2_+_6253246 | 2.52 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr19_-_5103141 | 2.22 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr8_+_21353878 | 2.20 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr8_-_14126646 | 2.14 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr2_+_33326522 | 2.13 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr6_+_50381665 | 2.02 |
ENSDART00000141128
|
cyc1
|
cytochrome c-1 |
| chr9_-_21918963 | 2.00 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr10_-_7756865 | 1.94 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr24_-_40744672 | 1.92 |
ENSDART00000160672
|
CU633479.1
|
|
| chr21_+_25777425 | 1.84 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr21_-_11654422 | 1.84 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr6_-_3982783 | 1.72 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr17_+_16046314 | 1.70 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr19_+_2631565 | 1.67 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr20_-_23426339 | 1.67 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr16_+_47207691 | 1.61 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr11_-_1550709 | 1.61 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr1_-_18811517 | 1.60 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr12_+_24952902 | 1.58 |
ENSDART00000189086
ENSDART00000014868 |
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr21_+_34088377 | 1.57 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr17_+_16046132 | 1.57 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr22_-_15593824 | 1.55 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr8_+_45334255 | 1.55 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr23_-_31913069 | 1.53 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr24_+_16547035 | 1.51 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr20_-_20931197 | 1.50 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr12_-_14143344 | 1.49 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr21_-_39177564 | 1.49 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr19_-_32710922 | 1.45 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr21_-_32060993 | 1.43 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr5_+_35786141 | 1.41 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr22_+_16535575 | 1.41 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr7_-_71389375 | 1.40 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr25_+_3306620 | 1.38 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr3_-_30685401 | 1.36 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr20_+_52554352 | 1.35 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr10_-_21362320 | 1.32 |
ENSDART00000189789
|
avd
|
avidin |
| chr23_+_11285662 | 1.31 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr18_-_40708537 | 1.30 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr23_+_31913292 | 1.29 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr18_+_24921587 | 1.29 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr10_-_21362071 | 1.25 |
ENSDART00000125167
|
avd
|
avidin |
| chr21_+_34088110 | 1.25 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr16_-_29387215 | 1.20 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr15_-_9272328 | 1.19 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr18_+_19456648 | 1.17 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr23_+_44741500 | 1.16 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr14_+_22113331 | 1.13 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr23_-_31913231 | 1.12 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr15_+_1534644 | 1.10 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr21_-_40174647 | 1.10 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr24_+_12835935 | 1.10 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr7_+_23292133 | 1.09 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr10_-_25217347 | 1.07 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr23_+_31912882 | 1.06 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr5_+_52167986 | 1.06 |
ENSDART00000162256
ENSDART00000073626 |
slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr7_+_20503344 | 1.05 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr1_+_8694196 | 1.03 |
ENSDART00000025604
|
zgc:77849
|
zgc:77849 |
| chr20_-_20930926 | 1.03 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr6_+_21001264 | 1.02 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr15_-_43284021 | 1.02 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr6_-_14292307 | 1.01 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr6_+_50381347 | 1.00 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr5_+_37903790 | 0.99 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr16_-_42056137 | 0.95 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr16_-_17347727 | 0.95 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr25_+_7504314 | 0.93 |
ENSDART00000163231
|
ifitm5
|
interferon induced transmembrane protein 5 |
| chr21_+_15592426 | 0.93 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr22_+_18477934 | 0.90 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr13_+_38430466 | 0.90 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr16_+_29509133 | 0.89 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr23_-_25135046 | 0.88 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr12_+_10631266 | 0.85 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr22_-_19552796 | 0.84 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr17_+_23298928 | 0.83 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr22_+_4488454 | 0.81 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr3_-_16719244 | 0.80 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr5_-_68093169 | 0.76 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr22_-_2937503 | 0.75 |
ENSDART00000092991
ENSDART00000131110 |
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr22_-_20166660 | 0.75 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr20_-_52338782 | 0.75 |
ENSDART00000109735
ENSDART00000132941 |
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr9_+_29548195 | 0.74 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr24_+_1023839 | 0.73 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr6_-_12172424 | 0.71 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr13_+_24750078 | 0.69 |
ENSDART00000021053
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr8_+_50953776 | 0.68 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr14_-_22113600 | 0.68 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr8_+_7144066 | 0.68 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr1_-_55248496 | 0.67 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr20_-_14114078 | 0.67 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr17_-_16422654 | 0.66 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr3_-_39695856 | 0.66 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr21_-_40173821 | 0.66 |
ENSDART00000180667
|
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr7_+_56098590 | 0.66 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr24_+_21540842 | 0.65 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr11_-_30634286 | 0.64 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr24_+_40860320 | 0.64 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr7_-_51773166 | 0.64 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr1_+_35985813 | 0.63 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr9_-_31278048 | 0.63 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr5_+_36611128 | 0.63 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr11_-_35171162 | 0.62 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr8_+_49117518 | 0.62 |
ENSDART00000079631
|
rad21l1
|
RAD21 cohesin complex component like 1 |
| chr15_-_23376541 | 0.61 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr8_+_2757821 | 0.61 |
ENSDART00000051403
ENSDART00000160551 |
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr7_+_26649319 | 0.61 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr4_-_5455506 | 0.61 |
ENSDART00000156593
ENSDART00000154676 |
si:dkey-14d8.22
|
si:dkey-14d8.22 |
| chr24_+_19415124 | 0.60 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr21_-_35419486 | 0.59 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr1_+_52929185 | 0.58 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr6_-_50203682 | 0.57 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr14_+_45406299 | 0.56 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr10_+_6884627 | 0.55 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr13_+_35637048 | 0.53 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr12_-_30359498 | 0.51 |
ENSDART00000152981
ENSDART00000189988 |
tdrd1
|
tudor domain containing 1 |
| chr1_+_18811679 | 0.48 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr3_-_30488063 | 0.48 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr3_-_39696066 | 0.48 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr14_-_33105434 | 0.48 |
ENSDART00000163795
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr11_+_29770966 | 0.47 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr21_+_13366353 | 0.47 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr16_+_13883872 | 0.46 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr1_-_26444075 | 0.46 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr3_+_26244353 | 0.45 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr5_+_4436405 | 0.45 |
ENSDART00000167969
|
CABZ01079241.1
|
|
| chr1_-_29139141 | 0.44 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr11_+_38280454 | 0.43 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr6_+_102506 | 0.42 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr19_-_30524952 | 0.42 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr24_-_37640705 | 0.42 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr11_+_33818179 | 0.42 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr8_+_36500308 | 0.41 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr11_-_41220794 | 0.39 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr10_+_15777258 | 0.38 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr21_+_44857293 | 0.38 |
ENSDART00000134365
ENSDART00000168217 ENSDART00000065083 |
fstl4
|
follistatin-like 4 |
| chr21_-_22827548 | 0.38 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr17_-_20118145 | 0.38 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr16_+_5774977 | 0.37 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr2_+_50608099 | 0.37 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chrM_+_3803 | 0.37 |
ENSDART00000093596
|
mt-nd1
|
NADH dehydrogenase 1, mitochondrial |
| chr14_+_33329761 | 0.36 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr11_-_7261717 | 0.36 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr20_-_45661049 | 0.36 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr15_+_36309070 | 0.36 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr24_-_6078222 | 0.34 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr13_+_25486608 | 0.34 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr17_-_31695217 | 0.34 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr8_+_41037541 | 0.33 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr20_-_16171297 | 0.33 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr19_-_20113696 | 0.33 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr24_+_9412450 | 0.33 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr17_-_31639845 | 0.33 |
ENSDART00000154196
|
znf839
|
zinc finger protein 839 |
| chr20_-_32270866 | 0.32 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr22_-_12160283 | 0.30 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr7_+_28612671 | 0.30 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr16_+_21330634 | 0.30 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr10_+_6884123 | 0.30 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr25_+_11456696 | 0.29 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr4_+_9400012 | 0.29 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr19_+_43297546 | 0.29 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr3_+_45364849 | 0.28 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr11_+_31864921 | 0.28 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr2_-_32384683 | 0.28 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr14_-_8940499 | 0.28 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr2_-_30668580 | 0.28 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr23_+_40460333 | 0.27 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr25_-_2723682 | 0.27 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr9_+_25776971 | 0.27 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr9_-_746317 | 0.27 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr16_-_43344859 | 0.25 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr19_-_19379084 | 0.25 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr15_-_34785594 | 0.24 |
ENSDART00000154256
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr24_-_24724233 | 0.24 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr19_+_10339538 | 0.24 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr19_+_40069524 | 0.23 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chrM_+_8130 | 0.22 |
ENSDART00000093609
|
mt-co2
|
cytochrome c oxidase II, mitochondrial |
| chr21_+_42226113 | 0.22 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr12_+_2446837 | 0.22 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr11_-_30158191 | 0.22 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr20_-_45062514 | 0.22 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr22_-_10121880 | 0.21 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr1_+_21731382 | 0.21 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr7_+_34592526 | 0.21 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr7_-_72261721 | 0.20 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr14_+_6962271 | 0.20 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr4_+_9177997 | 0.20 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr10_+_43039947 | 0.20 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr8_-_25033681 | 0.19 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr12_-_4408828 | 0.18 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr8_-_19216657 | 0.18 |
ENSDART00000135096
ENSDART00000135869 ENSDART00000145951 |
si:ch73-222f22.2
si:ch73-222f22.2
|
si:ch73-222f22.2 si:ch73-222f22.2 |
| chr5_+_28770273 | 0.17 |
ENSDART00000114473
|
trafd1
|
TRAF-type zinc finger domain containing 1 |
| chr18_-_29896367 | 0.16 |
ENSDART00000191303
|
cmc2
|
C-x(9)-C motif containing 2 |
| chr8_-_16406440 | 0.16 |
ENSDART00000034004
|
faf1
|
Fas (TNFRSF6) associated factor 1 |
| chr19_-_30510259 | 0.15 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr3_+_33341640 | 0.15 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr22_-_14739491 | 0.15 |
ENSDART00000133385
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr21_-_36453594 | 0.14 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mixl1+mxtx1+sebox
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.6 | 2.2 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.5 | 1.4 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 1.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 1.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 3.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.3 | 1.8 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.3 | 1.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 0.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 3.5 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.8 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 1.5 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 0.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 1.7 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 0.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.6 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.2 | 8.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 0.8 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) C4-dicarboxylate transport(GO:0015740) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 2.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 1.2 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 1.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 7.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.9 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.3 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 0.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.8 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.3 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 0.3 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 1.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.6 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 3.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 0.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.4 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 1.6 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.6 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.2 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 2.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.7 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.9 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.2 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.0 | 0.3 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.0 | 2.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.5 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.2 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 1.5 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0060114 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.7 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 2.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.5 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.5 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.0 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.0 | 1.2 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 1.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.1 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.0 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 1.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 8.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.7 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 0.8 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 0.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.0 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.8 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 1.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.6 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.5 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.5 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.3 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 4.1 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.6 | 2.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.5 | 2.6 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 2.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 1.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 2.5 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 1.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 0.7 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 1.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.2 | 0.9 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 7.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.8 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 1.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.5 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 1.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.6 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.4 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.8 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.2 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 3.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.1 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.0 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.9 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 2.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 5.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |