Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
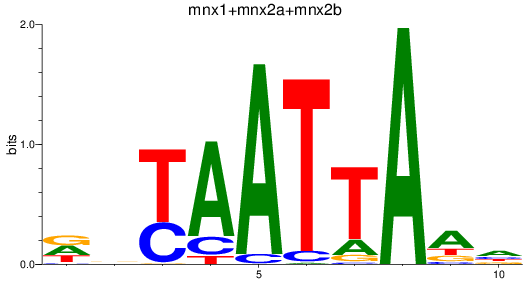
Results for mnx1+mnx2a+mnx2b
Z-value: 0.59
Transcription factors associated with mnx1+mnx2a+mnx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mnx2b
|
ENSDARG00000030350 | motor neuron and pancreas homeobox 2b |
|
mnx1
|
ENSDARG00000035984 | motor neuron and pancreas homeobox 1 |
|
mnx2a
|
ENSDARG00000042106 | motor neuron and pancreas homeobox 2a |
|
mnx1
|
ENSDARG00000109419 | motor neuron and pancreas homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mnx1 | dr11_v1_chr7_+_40638210_40638210 | -0.12 | 2.7e-01 | Click! |
| mnx2a | dr11_v1_chr9_+_7724152_7724152 | -0.12 | 2.7e-01 | Click! |
| mnx2b | dr11_v1_chr1_-_5455498_5455502 | -0.11 | 2.7e-01 | Click! |
Activity profile of mnx1+mnx2a+mnx2b motif
Sorted Z-values of mnx1+mnx2a+mnx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_21867307 | 5.45 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr13_-_36034582 | 4.80 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr20_-_40755614 | 4.58 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr17_+_43867889 | 4.06 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr18_+_15644559 | 3.02 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr6_+_45918981 | 2.88 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr7_-_26306546 | 2.81 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr22_-_24818066 | 2.61 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr22_+_19552987 | 2.52 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr11_-_19775182 | 2.44 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr16_-_42965192 | 2.33 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr9_+_38163876 | 2.31 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr13_-_8692860 | 2.24 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr22_-_36836085 | 2.22 |
ENSDART00000131451
|
cart1
|
cocaine- and amphetamine-regulated transcript 1 |
| chr4_-_9891874 | 2.20 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr8_-_30979494 | 2.16 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr6_-_43283122 | 2.15 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr18_-_14677936 | 2.15 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr25_+_10410620 | 2.01 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr10_-_8060573 | 1.92 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr8_+_21229718 | 1.86 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr10_-_24371312 | 1.85 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr20_-_34028967 | 1.81 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr16_+_26777473 | 1.78 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr15_+_6109861 | 1.73 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr6_-_7776612 | 1.72 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr15_+_21262917 | 1.71 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr2_-_33993533 | 1.68 |
ENSDART00000140910
ENSDART00000077304 |
ptch2
|
patched 2 |
| chr2_-_28671139 | 1.65 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr17_+_28103675 | 1.63 |
ENSDART00000188078
|
zgc:91908
|
zgc:91908 |
| chr24_-_30862168 | 1.62 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr3_-_27647845 | 1.61 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr21_-_25722834 | 1.55 |
ENSDART00000101208
|
abhd11
|
abhydrolase domain containing 11 |
| chr10_+_7593185 | 1.48 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr24_-_29963858 | 1.47 |
ENSDART00000183442
|
CR352310.1
|
|
| chr17_-_40956035 | 1.46 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr17_-_32863250 | 1.44 |
ENSDART00000167292
|
prox1a
|
prospero homeobox 1a |
| chr3_+_32365811 | 1.44 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr7_+_69019851 | 1.41 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr1_+_44173245 | 1.39 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr18_+_8320165 | 1.39 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr4_+_16885854 | 1.38 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr2_+_41526904 | 1.33 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr9_+_34641237 | 1.32 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr16_+_39159752 | 1.32 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr2_-_38363017 | 1.32 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr9_-_20372977 | 1.30 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr8_+_39998467 | 1.28 |
ENSDART00000073782
ENSDART00000134452 |
ggt5a
|
gamma-glutamyltransferase 5a |
| chr16_+_42471455 | 1.27 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr12_+_30367371 | 1.24 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr22_-_20166660 | 1.23 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr13_+_13945218 | 1.23 |
ENSDART00000089501
ENSDART00000142997 |
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr12_-_35830625 | 1.23 |
ENSDART00000180028
|
CU459056.1
|
|
| chr20_-_37813863 | 1.18 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr11_-_37691449 | 1.17 |
ENSDART00000185340
|
zgc:158258
|
zgc:158258 |
| chr18_-_2433011 | 1.16 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr14_+_35405518 | 1.16 |
ENSDART00000171565
|
zbtb3
|
zinc finger and BTB domain containing 3 |
| chr14_-_22113600 | 1.13 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr14_+_16151368 | 1.13 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr20_-_19511700 | 1.08 |
ENSDART00000040191
|
snx17
|
sorting nexin 17 |
| chr2_-_40135942 | 1.08 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr5_+_37032038 | 1.07 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr10_+_29850330 | 1.05 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr3_+_32832538 | 1.05 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr13_-_31017960 | 1.04 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr7_-_31321027 | 1.04 |
ENSDART00000186878
|
CR356242.1
|
|
| chr16_-_17586883 | 1.03 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr22_-_10440688 | 1.03 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr1_+_10318089 | 1.01 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr3_-_16719244 | 1.00 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr17_+_16046314 | 0.99 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr1_-_50859053 | 0.98 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr17_-_21162821 | 0.97 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr8_-_24252933 | 0.97 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr21_-_7035599 | 0.96 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr14_+_23717165 | 0.95 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr15_-_16177603 | 0.94 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr19_+_30884706 | 0.94 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr16_+_33902006 | 0.93 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr23_+_28809002 | 0.93 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr20_+_27712714 | 0.92 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr1_+_52392511 | 0.90 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr17_+_14965570 | 0.89 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr3_+_45368973 | 0.86 |
ENSDART00000187282
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr22_-_10397600 | 0.86 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr14_+_35428152 | 0.86 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr3_-_26806032 | 0.86 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr5_+_66433287 | 0.86 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr17_+_16090436 | 0.85 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr22_+_10440991 | 0.82 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr6_+_3280939 | 0.82 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr12_+_30367079 | 0.82 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr16_+_23087326 | 0.82 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr8_-_25034411 | 0.81 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr15_+_25528290 | 0.81 |
ENSDART00000123143
|
npat
|
nuclear protein, ataxia-telangiectasia locus |
| chr14_+_16151636 | 0.80 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr3_-_38783951 | 0.78 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr24_-_35561672 | 0.78 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr10_-_8046764 | 0.77 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr13_-_25819825 | 0.77 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr9_-_24244383 | 0.76 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr7_+_17229980 | 0.75 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr10_+_33744098 | 0.74 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr16_-_17162843 | 0.73 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr7_-_71389375 | 0.73 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr19_+_42432625 | 0.72 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr11_-_44801968 | 0.71 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr19_-_30524952 | 0.71 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr1_+_45056371 | 0.71 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr5_-_25733745 | 0.71 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr22_-_7129631 | 0.70 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr11_-_39118882 | 0.69 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr1_+_12766351 | 0.66 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr19_-_19379084 | 0.65 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr1_-_45616470 | 0.65 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr24_-_40860603 | 0.64 |
ENSDART00000188032
|
CU633479.7
|
|
| chr5_+_25733774 | 0.63 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr10_+_42521943 | 0.63 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr3_+_32553714 | 0.63 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr4_-_15103646 | 0.63 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr2_-_38206034 | 0.62 |
ENSDART00000144518
ENSDART00000137395 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr16_+_35916371 | 0.61 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr25_-_27722614 | 0.61 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr19_+_40069524 | 0.60 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr23_-_40766518 | 0.60 |
ENSDART00000127420
|
si:dkeyp-27c8.2
|
si:dkeyp-27c8.2 |
| chr15_+_857148 | 0.59 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr17_+_31221761 | 0.58 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr8_-_39822917 | 0.57 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr20_-_48485354 | 0.57 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr15_-_26931541 | 0.57 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr21_+_5580948 | 0.56 |
ENSDART00000160373
|
ly6m7
|
lymphocyte antigen 6 family member M7 |
| chr8_+_2487250 | 0.56 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr17_+_49500820 | 0.55 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr8_+_23355484 | 0.53 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr18_+_19456648 | 0.52 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr10_+_11282883 | 0.52 |
ENSDART00000135355
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr7_+_25059845 | 0.50 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr3_+_13624815 | 0.49 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr10_-_8053385 | 0.49 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr17_-_44249720 | 0.48 |
ENSDART00000156648
|
otx2b
|
orthodenticle homeobox 2b |
| chr19_-_5103141 | 0.48 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr5_+_66353750 | 0.48 |
ENSDART00000143410
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr1_-_44701313 | 0.48 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr12_+_24342303 | 0.47 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr20_-_45812144 | 0.46 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr14_+_33329420 | 0.45 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr19_-_44065581 | 0.43 |
ENSDART00000006338
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr10_-_25217347 | 0.42 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr10_+_43039947 | 0.42 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr12_+_22580579 | 0.42 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr7_+_19552381 | 0.40 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr2_-_32384683 | 0.40 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr13_-_4018888 | 0.40 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr5_+_66353589 | 0.40 |
ENSDART00000138246
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr20_+_31076488 | 0.40 |
ENSDART00000136255
ENSDART00000008840 |
otofa
|
otoferlin a |
| chr14_+_49135264 | 0.39 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr15_+_5360407 | 0.39 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr17_+_23298928 | 0.38 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr13_-_27354003 | 0.37 |
ENSDART00000101479
ENSDART00000044652 |
ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr16_-_42056137 | 0.36 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr18_-_33979693 | 0.36 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr23_+_30967686 | 0.36 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr1_-_10647484 | 0.35 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr8_-_50888806 | 0.34 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr11_-_6452444 | 0.34 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr25_+_35553542 | 0.34 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr6_-_39649504 | 0.32 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr8_+_2487883 | 0.31 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr1_+_50613868 | 0.31 |
ENSDART00000111114
|
HERC5
|
si:ch73-190m4.1 |
| chr24_+_5912635 | 0.31 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr5_+_66063807 | 0.31 |
ENSDART00000122405
|
jak2b
|
Janus kinase 2b |
| chr24_-_6078222 | 0.31 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr8_+_45334255 | 0.30 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr11_+_24820542 | 0.30 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr20_-_7128612 | 0.30 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr3_+_46635527 | 0.28 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr5_-_48664522 | 0.28 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr23_-_36418059 | 0.27 |
ENSDART00000135232
|
znf740b
|
zinc finger protein 740b |
| chr20_-_23426339 | 0.27 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr9_+_50001746 | 0.26 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr3_-_32873641 | 0.25 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr10_-_28028998 | 0.25 |
ENSDART00000023545
ENSDART00000143487 |
ints2
|
integrator complex subunit 2 |
| chr19_+_22062202 | 0.25 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr1_-_22512063 | 0.24 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr4_-_16334362 | 0.23 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr1_+_11977426 | 0.22 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr9_-_14273652 | 0.21 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr1_+_33969015 | 0.20 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr8_-_34052019 | 0.19 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr23_+_45200481 | 0.19 |
ENSDART00000004357
ENSDART00000111126 ENSDART00000193560 ENSDART00000190476 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr16_-_29387215 | 0.19 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr2_+_20332044 | 0.19 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr20_-_35040041 | 0.19 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr6_-_11780070 | 0.18 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr3_+_33341640 | 0.18 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr13_+_38430466 | 0.17 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr4_+_12612145 | 0.17 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr1_-_32111882 | 0.17 |
ENSDART00000112639
ENSDART00000101958 |
pnpla4
|
patatin-like phospholipase domain containing 4 |
| chr5_+_20147830 | 0.17 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr11_+_18157260 | 0.17 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mnx1+mnx2a+mnx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 1.3 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.3 | 1.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 1.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.3 | 2.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 1.3 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.3 | 1.0 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 2.9 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 0.9 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.2 | 3.0 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 0.9 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.2 | 0.6 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 1.0 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.2 | 1.7 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 1.1 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.2 | 0.5 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 1.1 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.1 | 1.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.9 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.8 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.5 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.8 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.4 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.6 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.7 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.1 | 0.9 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.6 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.9 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 1.6 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 1.4 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 2.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 0.5 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 0.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 2.3 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.8 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.7 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 1.4 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 3.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.8 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 1.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.8 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 2.8 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 1.3 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 1.0 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 2.1 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 2.0 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.0 | 1.9 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.4 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.5 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 1.0 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 1.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.3 | 0.9 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 2.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.3 | 1.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 1.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 1.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 1.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 3.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 3.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 3.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.8 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 2.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 2.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.5 | 3.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.4 | 1.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.4 | 1.7 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.4 | 2.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.3 | 2.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 1.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 1.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 2.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.9 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.2 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 1.3 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.4 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.0 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.5 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 3.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.0 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 2.5 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 2.1 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 1.7 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 2.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.2 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 1.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |