Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
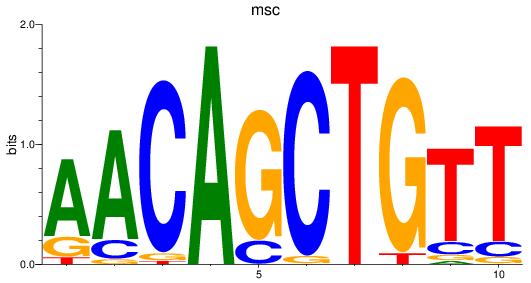
Results for msc
Z-value: 0.83
Transcription factors associated with msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msc
|
ENSDARG00000110016 | musculin (activated B-cell factor-1) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msc | dr11_v1_chr24_+_13735616_13735616 | -0.38 | 1.4e-04 | Click! |
Activity profile of msc motif
Sorted Z-values of msc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_18057851 | 6.61 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr21_+_27382893 | 6.43 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr23_-_18057553 | 6.43 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr20_-_27325258 | 6.35 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr13_+_24834199 | 5.90 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr21_-_43952958 | 5.36 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr5_-_51819027 | 5.21 |
ENSDART00000164267
|
homer1b
|
homer scaffolding protein 1b |
| chr6_+_13742899 | 5.02 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr1_-_25911292 | 4.90 |
ENSDART00000145012
|
usp53b
|
ubiquitin specific peptidase 53b |
| chr15_-_15357178 | 4.72 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr5_-_38451082 | 4.55 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr6_+_23887314 | 4.39 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr1_+_25801648 | 4.38 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr9_+_40939336 | 4.34 |
ENSDART00000100386
|
mstnb
|
myostatin b |
| chr2_-_44255537 | 4.22 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr5_-_31904562 | 4.07 |
ENSDART00000140640
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr23_-_18057270 | 3.96 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr7_+_31879649 | 3.90 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr11_-_5865744 | 3.83 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr7_+_26224211 | 3.81 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr14_-_32255126 | 3.74 |
ENSDART00000017259
|
fgf13a
|
fibroblast growth factor 13a |
| chr7_+_6969909 | 3.69 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr7_+_31879986 | 3.67 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr13_-_41155472 | 3.60 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr11_-_1291012 | 3.56 |
ENSDART00000158390
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr13_-_31452516 | 3.45 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr2_-_42128714 | 3.34 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr1_+_33401774 | 3.29 |
ENSDART00000149786
ENSDART00000110026 ENSDART00000148946 |
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr10_+_15777064 | 3.26 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr3_-_28075756 | 3.20 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr10_+_15777258 | 3.19 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr7_+_7048245 | 3.19 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr20_+_35382482 | 3.19 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr5_+_3501859 | 3.15 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr5_+_42097608 | 3.08 |
ENSDART00000014404
|
p2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr7_-_23745984 | 3.08 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr15_+_40188076 | 3.02 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr19_-_9711472 | 2.93 |
ENSDART00000016197
ENSDART00000175075 |
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr21_+_11468642 | 2.92 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr6_+_39222598 | 2.90 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr9_-_49531762 | 2.85 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr2_-_32551178 | 2.85 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr19_-_10425140 | 2.76 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr7_+_19552381 | 2.70 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr1_-_54997746 | 2.60 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr14_+_21783229 | 2.59 |
ENSDART00000170784
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr23_+_35714574 | 2.55 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr6_+_40587551 | 2.54 |
ENSDART00000155554
|
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr13_-_40499296 | 2.53 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr15_+_33989181 | 2.51 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr3_-_36115339 | 2.47 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr24_+_26276805 | 2.45 |
ENSDART00000089749
|
adipoqa
|
adiponectin, C1Q and collagen domain containing, a |
| chr21_+_11468934 | 2.45 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr22_+_16308450 | 2.44 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr21_-_16632808 | 2.43 |
ENSDART00000172645
|
unc5da
|
unc-5 netrin receptor Da |
| chr18_-_5781922 | 2.41 |
ENSDART00000128722
|
RGS9BP
|
si:ch73-167i17.6 |
| chr23_-_8373676 | 2.38 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr11_-_17713987 | 2.37 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr14_+_5936996 | 2.37 |
ENSDART00000097144
ENSDART00000126777 |
kctd8
|
potassium channel tetramerization domain containing 8 |
| chr24_+_38155830 | 2.35 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr15_-_9421481 | 2.33 |
ENSDART00000189045
ENSDART00000177158 |
sacs
|
sacsin molecular chaperone |
| chr9_-_98982 | 2.31 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr10_+_34315719 | 2.30 |
ENSDART00000135303
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr22_+_16308806 | 2.30 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr23_-_32162810 | 2.25 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr10_-_34741738 | 2.24 |
ENSDART00000163072
|
dclk1a
|
doublecortin-like kinase 1a |
| chr7_+_27277236 | 2.23 |
ENSDART00000185161
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr17_-_14836320 | 2.22 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr10_-_25217347 | 2.18 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr6_-_8865217 | 2.16 |
ENSDART00000151443
|
tmeff2b
|
transmembrane protein with EGF-like and two follistatin-like domains 2b |
| chr5_-_19400166 | 2.15 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr17_-_14726824 | 2.11 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr14_+_21783400 | 2.11 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr7_+_20471315 | 2.11 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr6_-_16667886 | 2.10 |
ENSDART00000180854
ENSDART00000190116 |
unc80
|
unc-80 homolog (C. elegans) |
| chr13_+_1575276 | 2.07 |
ENSDART00000165987
|
DST
|
dystonin |
| chr19_-_10432134 | 2.06 |
ENSDART00000081440
|
il11b
|
interleukin 11b |
| chr24_+_20575259 | 2.03 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr19_+_1184878 | 2.03 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr22_+_465269 | 2.01 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr23_+_29908348 | 2.01 |
ENSDART00000186622
ENSDART00000150174 |
si:ch73-236e11.2
|
si:ch73-236e11.2 |
| chr24_+_24461558 | 2.00 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr11_+_14622379 | 1.98 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr17_-_4245311 | 1.96 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr15_+_9327252 | 1.94 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr8_-_44004135 | 1.92 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr20_-_39391833 | 1.91 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr19_+_9174166 | 1.91 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr2_+_45696743 | 1.90 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
|
| chr9_+_52492639 | 1.90 |
ENSDART00000078939
|
march4
|
membrane-associated ring finger (C3HC4) 4 |
| chr25_+_36045072 | 1.89 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr3_+_29714775 | 1.89 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr5_+_57924611 | 1.88 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr15_-_32383529 | 1.88 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr19_-_33087246 | 1.86 |
ENSDART00000052078
|
klhl38a
|
kelch-like family member 38a |
| chr20_-_42439896 | 1.85 |
ENSDART00000061049
|
vgll2a
|
vestigial-like family member 2a |
| chr6_-_10912424 | 1.85 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr22_-_10470663 | 1.84 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr8_-_43997538 | 1.81 |
ENSDART00000186449
|
rimbp2
|
RIMS binding protein 2 |
| chr8_-_11229523 | 1.77 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr6_+_27339962 | 1.71 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr12_-_28570989 | 1.63 |
ENSDART00000008010
|
pdk2a
|
pyruvate dehydrogenase kinase, isozyme 2a |
| chr1_-_17650223 | 1.60 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr23_-_30954738 | 1.60 |
ENSDART00000188996
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr14_-_9355177 | 1.58 |
ENSDART00000138535
|
fam46d
|
family with sequence similarity 46, member D |
| chr19_-_32888758 | 1.57 |
ENSDART00000052080
|
laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr18_-_7948188 | 1.54 |
ENSDART00000091805
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr2_-_15040345 | 1.52 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr2_-_23768818 | 1.52 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr1_-_14258409 | 1.50 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr21_-_17482465 | 1.50 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr7_-_35516251 | 1.49 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr2_-_36933472 | 1.48 |
ENSDART00000170405
|
BX470229.2
|
|
| chr20_+_27020201 | 1.48 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr5_-_60159116 | 1.48 |
ENSDART00000147675
|
si:dkey-280e8.1
|
si:dkey-280e8.1 |
| chr10_-_25069155 | 1.46 |
ENSDART00000078226
ENSDART00000181941 |
mtnr1bb
|
melatonin receptor 1Bb |
| chr4_+_12617108 | 1.46 |
ENSDART00000134362
ENSDART00000112860 |
lmo3
|
LIM domain only 3 |
| chr4_+_12615836 | 1.46 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr13_+_24287093 | 1.42 |
ENSDART00000058628
|
ccsapb
|
centriole, cilia and spindle-associated protein b |
| chr6_+_4872883 | 1.41 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr17_+_6276559 | 1.41 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr3_+_20156956 | 1.40 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr4_-_20081621 | 1.40 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr20_+_18580176 | 1.40 |
ENSDART00000185310
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr1_+_2101541 | 1.39 |
ENSDART00000128187
ENSDART00000167050 ENSDART00000182153 ENSDART00000122626 ENSDART00000164488 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr1_+_20593653 | 1.38 |
ENSDART00000132440
|
si:ch211-142c4.1
|
si:ch211-142c4.1 |
| chr23_-_27647769 | 1.38 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr5_-_10768258 | 1.37 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr6_+_46259950 | 1.34 |
ENSDART00000032326
|
zgc:162324
|
zgc:162324 |
| chr18_+_39028417 | 1.34 |
ENSDART00000148428
|
myo5aa
|
myosin VAa |
| chr19_-_19505167 | 1.33 |
ENSDART00000160582
|
creb5a
|
cAMP responsive element binding protein 5a |
| chr7_+_27253063 | 1.32 |
ENSDART00000191138
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr12_+_33038757 | 1.32 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr1_-_29747702 | 1.31 |
ENSDART00000133225
ENSDART00000189670 |
spp2
|
secreted phosphoprotein 2 |
| chr20_-_39391492 | 1.30 |
ENSDART00000184886
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr4_+_12113105 | 1.29 |
ENSDART00000182399
|
tmem178b
|
transmembrane protein 178B |
| chr7_+_66634167 | 1.29 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr17_+_12075805 | 1.28 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr20_-_18915376 | 1.27 |
ENSDART00000063725
|
xkr6b
|
XK, Kell blood group complex subunit-related family, member 6b |
| chr1_-_17587552 | 1.27 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr16_+_28728347 | 1.26 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr20_+_25645459 | 1.26 |
ENSDART00000143555
ENSDART00000036350 |
aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr11_-_36963988 | 1.26 |
ENSDART00000168288
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr2_-_17115256 | 1.26 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr8_-_24965308 | 1.26 |
ENSDART00000087293
ENSDART00000146805 |
kcnc4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr17_-_21066075 | 1.25 |
ENSDART00000078763
ENSDART00000104327 |
vsx1
|
visual system homeobox 1 homolog, chx10-like |
| chr21_-_27123139 | 1.23 |
ENSDART00000077478
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr18_+_19820008 | 1.22 |
ENSDART00000079691
ENSDART00000135049 |
iqch
|
IQ motif containing H |
| chr4_-_75175407 | 1.21 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr5_-_22019061 | 1.21 |
ENSDART00000113066
|
amer1
|
APC membrane recruitment protein 1 |
| chr3_+_17910569 | 1.21 |
ENSDART00000080946
|
ttc25
|
tetratricopeptide repeat domain 25 |
| chr2_+_10642047 | 1.20 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr2_+_38881165 | 1.19 |
ENSDART00000141850
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr2_-_30659222 | 1.18 |
ENSDART00000145405
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr15_+_32727848 | 1.17 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
| chr21_-_38618540 | 1.17 |
ENSDART00000036600
|
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr6_+_28051978 | 1.16 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr12_-_10505986 | 1.16 |
ENSDART00000152672
|
zgc:152977
|
zgc:152977 |
| chr6_-_12722360 | 1.15 |
ENSDART00000090174
|
dock9b
|
dedicator of cytokinesis 9b |
| chr4_-_9586713 | 1.15 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr22_-_18179214 | 1.15 |
ENSDART00000129576
|
si:ch211-125m10.6
|
si:ch211-125m10.6 |
| chr10_+_24468922 | 1.15 |
ENSDART00000008248
ENSDART00000183510 |
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr22_+_24770744 | 1.15 |
ENSDART00000142882
|
si:rp71-23d18.4
|
si:rp71-23d18.4 |
| chr10_+_35491216 | 1.15 |
ENSDART00000109705
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr3_+_36515376 | 1.14 |
ENSDART00000161652
|
si:dkeyp-72e1.9
|
si:dkeyp-72e1.9 |
| chr9_+_2020667 | 1.13 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr15_-_38009344 | 1.13 |
ENSDART00000157094
|
si:dkey-238d18.6
|
si:dkey-238d18.6 |
| chr20_-_18731268 | 1.13 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr22_+_35472653 | 1.13 |
ENSDART00000076424
ENSDART00000187204 |
tctex1d2
|
Tctex1 domain containing 2 |
| chr24_-_9300160 | 1.12 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr23_-_24263474 | 1.11 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr21_-_32684570 | 1.10 |
ENSDART00000162873
|
adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr14_+_36628131 | 1.09 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr8_+_25247245 | 1.09 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr14_+_7939398 | 1.08 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr2_+_29257942 | 1.07 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr15_-_32383340 | 1.06 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr7_+_27059330 | 1.06 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr12_+_17504559 | 1.06 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr8_+_29742237 | 1.06 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr8_+_45334255 | 1.04 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr20_+_46560258 | 1.04 |
ENSDART00000122115
|
si:ch211-153j24.3
|
si:ch211-153j24.3 |
| chr23_-_7826849 | 1.03 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr17_-_50020672 | 1.03 |
ENSDART00000188942
|
filip1a
|
filamin A interacting protein 1a |
| chr10_+_10738880 | 1.01 |
ENSDART00000004181
|
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr16_+_30933044 | 0.99 |
ENSDART00000184453
|
gstk2
|
glutathione S-transferase kappa 2 |
| chr11_-_11266882 | 0.99 |
ENSDART00000020256
|
lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr10_+_26871751 | 0.99 |
ENSDART00000089205
|
ehbp1l1b
|
EH domain binding protein 1-like 1b |
| chr5_+_34622320 | 0.95 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr25_-_35542739 | 0.95 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr19_+_44039849 | 0.95 |
ENSDART00000086040
|
lrrc14b
|
leucine rich repeat containing 14B |
| chr16_-_13281380 | 0.95 |
ENSDART00000103882
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr18_+_25003207 | 0.94 |
ENSDART00000099476
ENSDART00000132285 |
fam174b
|
family with sequence similarity 174, member B |
| chr3_-_23596532 | 0.93 |
ENSDART00000124921
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr14_+_7939216 | 0.93 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr15_-_8517376 | 0.93 |
ENSDART00000186289
|
npas1
|
neuronal PAS domain protein 1 |
| chr15_+_37992275 | 0.91 |
ENSDART00000154928
|
si:dkey-238d18.7
|
si:dkey-238d18.7 |
| chr20_+_23501535 | 0.91 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr13_-_31164749 | 0.90 |
ENSDART00000049180
|
mapk8a
|
mitogen-activated protein kinase 8a |
| chr25_-_36044583 | 0.90 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
Network of associatons between targets according to the STRING database.
First level regulatory network of msc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.6 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 1.4 | 4.2 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 1.3 | 3.8 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 1.1 | 4.4 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 1.1 | 4.3 | GO:1901862 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.7 | 5.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.7 | 2.0 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.5 | 1.5 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.4 | 1.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 1.3 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.4 | 7.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.4 | 2.0 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.4 | 2.7 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.4 | 1.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.4 | 4.9 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.4 | 2.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.4 | 6.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 3.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.4 | 1.4 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.3 | 2.8 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.3 | 2.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.3 | 2.0 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 3.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 0.8 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.3 | 1.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 3.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.3 | 2.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 1.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 1.2 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.2 | 4.6 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 2.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 2.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 3.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 2.4 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.2 | 2.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 2.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 0.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 1.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 1.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.2 | 0.8 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.2 | 0.8 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.1 | 0.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 1.0 | GO:0044539 | very long-chain fatty acid catabolic process(GO:0042760) long-chain fatty acid import(GO:0044539) |
| 0.1 | 5.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 3.9 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 1.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.8 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 1.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.7 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.3 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 1.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 1.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 1.2 | GO:0042044 | fluid transport(GO:0042044) |
| 0.1 | 0.9 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.4 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 2.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 2.0 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 5.4 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 0.6 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.1 | 3.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.7 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.7 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.9 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 1.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.4 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 1.2 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 0.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.3 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 2.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 2.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.8 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 1.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.6 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 2.8 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 0.8 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.4 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 1.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.4 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.1 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 1.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 2.6 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.8 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.9 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.6 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.0 | 0.2 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 1.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.6 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) |
| 0.0 | 1.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 2.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.0 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 3.7 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.0 | 3.1 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.9 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 3.3 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 1.5 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 3.9 | GO:0003015 | heart process(GO:0003015) |
| 0.0 | 0.2 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 0.2 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 1.5 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.8 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 1.2 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 1.9 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 4.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.0 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 1.2 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.7 | GO:0014032 | neural crest cell development(GO:0014032) |
| 0.0 | 2.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 4.8 | GO:0007507 | heart development(GO:0007507) |
| 0.0 | 0.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 6.8 | GO:0016567 | protein ubiquitination(GO:0016567) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 6.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 4.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.4 | 1.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.4 | 1.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.3 | 1.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 8.5 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 0.9 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 4.2 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 0.7 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.2 | 2.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 5.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.6 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.4 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 2.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 3.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 9.1 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 3.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 6.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 4.2 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 1.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 2.3 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 6.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 10.8 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 3.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 4.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.7 | 7.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 2.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 4.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 5.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 2.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 1.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 6.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 6.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 2.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.3 | 3.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 1.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 3.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 4.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 2.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.2 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 2.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 1.4 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.2 | 1.4 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.2 | 4.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 0.7 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.2 | 0.7 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.2 | 2.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 0.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 2.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 0.8 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.0 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.4 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 3.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 3.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 5.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 2.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.1 | 1.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 2.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.3 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.1 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 6.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 2.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 3.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.8 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 4.3 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.7 | GO:0005501 | retinoid binding(GO:0005501) isoprenoid binding(GO:0019840) |
| 0.0 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 4.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 14.2 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 3.8 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 1.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 2.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 2.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 4.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.5 | 4.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 3.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 1.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |