Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
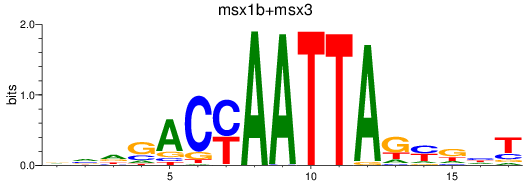
Results for msx1b+msx3
Z-value: 0.98
Transcription factors associated with msx1b+msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx1b
|
ENSDARG00000008886 | muscle segment homeobox 1b |
|
msx3
|
ENSDARG00000015674 | muscle segment homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx3 | dr11_v1_chr13_+_24662238_24662238 | -0.80 | 4.8e-22 | Click! |
| msx1b | dr11_v1_chr1_+_49352900_49352900 | -0.35 | 6.0e-04 | Click! |
Activity profile of msx1b+msx3 motif
Sorted Z-values of msx1b+msx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_29160102 | 15.54 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr15_-_5815006 | 11.00 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr24_+_38306010 | 10.75 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr13_+_22476742 | 10.45 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr9_-_22821901 | 8.30 |
ENSDART00000101711
|
neb
|
nebulin |
| chr18_-_16801033 | 8.00 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr18_+_9171778 | 7.65 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr5_+_56023186 | 7.44 |
ENSDART00000156230
|
fzd9a
|
frizzled class receptor 9a |
| chr1_-_25966068 | 7.27 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr20_+_46040666 | 7.09 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr12_+_28749189 | 6.96 |
ENSDART00000013980
|
tbx21
|
T-box 21 |
| chr24_+_20373605 | 6.96 |
ENSDART00000112349
|
plcd1a
|
phospholipase C, delta 1a |
| chr5_-_29534748 | 6.86 |
ENSDART00000159587
|
AL831768.1
|
|
| chr11_-_40457325 | 6.30 |
ENSDART00000128442
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr6_+_50451337 | 6.11 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr17_+_24851951 | 5.82 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr5_+_29794058 | 5.76 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr15_+_46344655 | 5.75 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr10_+_36178713 | 5.71 |
ENSDART00000140816
|
or108-3
|
odorant receptor, family D, subfamily 108, member 3 |
| chr3_+_23692462 | 5.59 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr6_-_19042294 | 5.50 |
ENSDART00000159461
|
si:rp71-81e14.2
|
si:rp71-81e14.2 |
| chr17_-_6514962 | 5.47 |
ENSDART00000163514
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr12_+_3078221 | 5.45 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr18_+_48423973 | 5.43 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr11_-_2478374 | 5.19 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr10_-_44560165 | 5.11 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr15_-_37733238 | 5.10 |
ENSDART00000115178
|
si:dkey-42l23.7
|
si:dkey-42l23.7 |
| chr20_-_40755614 | 5.08 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr12_-_33357655 | 5.07 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr19_-_8768564 | 5.04 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr15_-_37767901 | 4.93 |
ENSDART00000154763
|
si:dkey-42l23.4
|
si:dkey-42l23.4 |
| chr3_+_2719243 | 4.90 |
ENSDART00000189256
|
CR388047.3
|
|
| chr1_-_19402802 | 4.87 |
ENSDART00000135552
|
rbm47
|
RNA binding motif protein 47 |
| chr24_-_9979342 | 4.78 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr9_+_21277846 | 4.76 |
ENSDART00000139620
ENSDART00000110996 ENSDART00000111899 |
lats2
|
large tumor suppressor kinase 2 |
| chr11_+_37251825 | 4.75 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr4_+_77943184 | 4.59 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr19_-_19871211 | 4.58 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr16_+_33144306 | 4.31 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr1_+_40801696 | 4.30 |
ENSDART00000147497
|
cpz
|
carboxypeptidase Z |
| chr15_+_7057050 | 4.28 |
ENSDART00000061828
|
foxl2a
|
forkhead box L2a |
| chr5_+_27897504 | 4.26 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr15_-_25518084 | 4.26 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr16_-_41714988 | 4.16 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr16_+_33144112 | 4.09 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr11_+_31864921 | 4.06 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr24_-_25144441 | 4.03 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr22_+_12770877 | 4.01 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr16_-_35975254 | 3.97 |
ENSDART00000167537
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr1_+_54626491 | 3.95 |
ENSDART00000136063
|
si:ch211-202h22.9
|
si:ch211-202h22.9 |
| chr10_+_39212898 | 3.84 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr10_-_13343831 | 3.83 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr4_-_9891874 | 3.80 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr15_-_37709072 | 3.77 |
ENSDART00000157115
|
si:dkey-117a8.2
|
si:dkey-117a8.2 |
| chr15_-_37683768 | 3.76 |
ENSDART00000059609
|
si:dkey-117a8.3
|
si:dkey-117a8.3 |
| chr5_+_7564644 | 3.69 |
ENSDART00000192173
|
CABZ01039096.1
|
|
| chr7_+_8324506 | 3.66 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr15_-_37620167 | 3.65 |
ENSDART00000059582
|
si:ch211-137j23.8
|
si:ch211-137j23.8 |
| chr9_-_35633827 | 3.64 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr15_-_37719015 | 3.62 |
ENSDART00000154875
|
si:dkey-117a8.1
|
si:dkey-117a8.1 |
| chr23_+_16889352 | 3.62 |
ENSDART00000145275
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr17_+_26352372 | 3.54 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr11_-_25538341 | 3.53 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr22_+_5532003 | 3.48 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr19_-_18127808 | 3.41 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr15_-_37673733 | 3.39 |
ENSDART00000154513
|
si:dkey-117a8.4
|
si:dkey-117a8.4 |
| chr1_+_55730205 | 3.37 |
ENSDART00000148075
|
adgre10
|
adhesion G protein-coupled receptor E10 |
| chr25_+_6471401 | 3.36 |
ENSDART00000132927
|
snx33
|
sorting nexin 33 |
| chr8_-_27656765 | 3.34 |
ENSDART00000078491
|
mov10b.2
|
Moloney leukemia virus 10b, tandem duplicate 2 |
| chr5_-_47727819 | 3.33 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr10_+_36194527 | 3.10 |
ENSDART00000076810
|
or108-1
|
odorant receptor, family D, subfamily 108, member 1 |
| chr23_-_5729484 | 3.06 |
ENSDART00000000488
|
tnni1a
|
troponin I type 1a (skeletal, slow) |
| chr19_-_18127629 | 3.05 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr16_+_35595312 | 3.00 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr3_+_32663865 | 2.99 |
ENSDART00000190240
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr17_-_50071748 | 2.99 |
ENSDART00000075188
|
zgc:113886
|
zgc:113886 |
| chr11_+_24716837 | 2.98 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr1_+_29068654 | 2.97 |
ENSDART00000053932
|
cbsa
|
cystathionine-beta-synthase a |
| chr13_+_27328098 | 2.92 |
ENSDART00000037585
|
mb21d1
|
Mab-21 domain containing 1 |
| chr16_+_41171949 | 2.90 |
ENSDART00000135294
|
nek11
|
NIMA-related kinase 11 |
| chr13_+_15701596 | 2.66 |
ENSDART00000130832
|
trmt61a
|
tRNA methyltransferase 61A |
| chr8_-_53680653 | 2.59 |
ENSDART00000164739
|
wnt5a
|
wingless-type MMTV integration site family, member 5a |
| chr20_-_9095105 | 2.53 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr21_-_8085635 | 2.47 |
ENSDART00000082790
|
si:dkey-163m14.2
|
si:dkey-163m14.2 |
| chr10_-_40416054 | 2.45 |
ENSDART00000137947
ENSDART00000143606 |
taar20j
|
trace amine associated receptor 20j |
| chr21_-_22647695 | 2.41 |
ENSDART00000187553
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr8_-_15129573 | 2.39 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr25_+_10947743 | 2.38 |
ENSDART00000073381
|
mhc1lfa
|
major histocompatibility complex class I LFA |
| chr4_-_12997348 | 2.37 |
ENSDART00000171451
|
si:dkey-6a5.3
|
si:dkey-6a5.3 |
| chr14_+_34490445 | 2.36 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr10_+_39212601 | 2.32 |
ENSDART00000168778
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr20_+_46897504 | 2.31 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr2_+_31804582 | 2.29 |
ENSDART00000086646
|
rnf182
|
ring finger protein 182 |
| chr13_+_9468535 | 2.23 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr4_+_62576422 | 2.23 |
ENSDART00000180679
|
si:ch211-79g12.2
|
si:ch211-79g12.2 |
| chr4_-_42408339 | 2.21 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr25_-_13558172 | 2.18 |
ENSDART00000139970
ENSDART00000133789 ENSDART00000144426 |
ano10b
|
anoctamin 10b |
| chr4_-_14162327 | 2.15 |
ENSDART00000138656
|
si:dkey-234l24.1
|
si:dkey-234l24.1 |
| chr6_-_9695294 | 2.15 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr16_+_9713850 | 2.13 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr17_+_7513673 | 2.11 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr4_-_75681004 | 2.11 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr2_+_30369116 | 2.10 |
ENSDART00000142137
|
pi15b
|
peptidase inhibitor 15b |
| chr4_-_20292821 | 2.08 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr16_-_26855936 | 2.01 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr17_+_15041647 | 2.00 |
ENSDART00000108999
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr16_-_7379328 | 1.99 |
ENSDART00000060447
|
med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr21_+_15592426 | 1.88 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr4_-_14173138 | 1.87 |
ENSDART00000142487
|
si:dkey-234l24.7
|
si:dkey-234l24.7 |
| chr14_-_1543834 | 1.86 |
ENSDART00000185876
|
CABZ01109480.1
|
|
| chr1_-_7975755 | 1.86 |
ENSDART00000141765
|
si:dkey-79f11.7
|
si:dkey-79f11.7 |
| chr21_-_22648007 | 1.85 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr18_+_50461981 | 1.81 |
ENSDART00000158761
|
CU896640.1
|
|
| chr14_+_36521005 | 1.77 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr5_-_67629263 | 1.74 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr9_+_18829360 | 1.73 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr6_+_39370587 | 1.73 |
ENSDART00000157165
ENSDART00000155079 |
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr2_+_30032303 | 1.68 |
ENSDART00000151841
|
rbm33b
|
RNA binding motif protein 33b |
| chr19_+_9212031 | 1.63 |
ENSDART00000052930
|
ndufv1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1 |
| chr17_-_6618574 | 1.62 |
ENSDART00000184486
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr5_+_4806851 | 1.57 |
ENSDART00000067599
|
angptl2a
|
angiopoietin-like 2a |
| chr24_-_7957581 | 1.53 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr17_-_41081806 | 1.45 |
ENSDART00000030622
|
rbks
|
ribokinase |
| chr2_+_6253246 | 1.45 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr13_+_31402067 | 1.37 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr17_+_24821627 | 1.28 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr2_+_30368800 | 1.28 |
ENSDART00000179564
|
pi15b
|
peptidase inhibitor 15b |
| chr23_-_35483163 | 1.27 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr15_+_27384798 | 1.25 |
ENSDART00000164887
|
tbx4
|
T-box 4 |
| chr10_+_21867307 | 1.24 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr5_+_9224051 | 1.18 |
ENSDART00000139265
|
si:ch211-12e13.12
|
si:ch211-12e13.12 |
| chr18_-_46380471 | 1.16 |
ENSDART00000144444
ENSDART00000086772 |
slc19a3b
|
solute carrier family 19 (thiamine transporter), member 3b |
| chr13_-_9045879 | 1.15 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr14_+_20918701 | 1.14 |
ENSDART00000144736
ENSDART00000148204 |
zgc:66433
|
zgc:66433 |
| chr4_+_54519511 | 1.12 |
ENSDART00000161653
|
znf974
|
zinc finger protein 974 |
| chr22_+_9114416 | 1.06 |
ENSDART00000190169
|
nlrp15
|
NACHT, LRR and PYD domains-containing protein 15 |
| chr7_-_69647988 | 1.06 |
ENSDART00000169943
|
LO018231.1
|
|
| chr20_+_25568694 | 0.97 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr13_+_35955562 | 0.91 |
ENSDART00000137377
|
si:ch211-67f13.7
|
si:ch211-67f13.7 |
| chr5_-_68093169 | 0.86 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr25_-_25550938 | 0.86 |
ENSDART00000150412
ENSDART00000103622 |
irf7
|
interferon regulatory factor 7 |
| chr1_+_40801448 | 0.81 |
ENSDART00000187394
|
cpz
|
carboxypeptidase Z |
| chr23_+_22658700 | 0.79 |
ENSDART00000192248
|
eno1a
|
enolase 1a, (alpha) |
| chr13_-_9159852 | 0.78 |
ENSDART00000170185
ENSDART00000158697 ENSDART00000143393 ENSDART00000164973 ENSDART00000159910 |
HTRA2 (1 of many)
|
si:dkey-112g5.16 |
| chr5_+_19933356 | 0.77 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr17_-_2834764 | 0.76 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr11_-_1935883 | 0.72 |
ENSDART00000172885
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr11_+_2855430 | 0.65 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr22_+_1615691 | 0.60 |
ENSDART00000171555
|
si:ch211-255f4.13
|
si:ch211-255f4.13 |
| chr2_-_20788698 | 0.58 |
ENSDART00000181823
|
CABZ01020455.1
|
|
| chr5_+_872299 | 0.56 |
ENSDART00000130042
|
fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr18_+_43891051 | 0.55 |
ENSDART00000024213
|
cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr24_-_4782052 | 0.54 |
ENSDART00000149911
|
agtr1b
|
angiotensin II receptor, type 1b |
| chr4_-_62714083 | 0.54 |
ENSDART00000188299
|
si:dkey-28k24.2
|
si:dkey-28k24.2 |
| chr13_+_9521629 | 0.53 |
ENSDART00000149870
ENSDART00000137666 |
si:dkey-19p15.4
|
si:dkey-19p15.4 |
| chr25_+_19666621 | 0.51 |
ENSDART00000112746
|
zgc:171459
|
zgc:171459 |
| chr15_+_31303355 | 0.50 |
ENSDART00000143423
|
or116-2
|
odorant receptor, family F, subfamily 116, member 2 |
| chr18_+_34225520 | 0.47 |
ENSDART00000126115
|
v2rl1
|
vomeronasal 2 receptor, l1 |
| chr23_-_16485190 | 0.41 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr24_+_22485710 | 0.39 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr4_-_67969695 | 0.38 |
ENSDART00000190016
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr14_+_6535426 | 0.35 |
ENSDART00000055961
|
thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chr3_-_31079186 | 0.35 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr13_-_9213207 | 0.33 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr4_+_19700308 | 0.31 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr17_+_41081977 | 0.29 |
ENSDART00000137091
ENSDART00000143714 |
babam2
|
BRISC and BRCA1 A complex member 2 |
| chr4_-_14185225 | 0.24 |
ENSDART00000147061
|
si:dkey-234l24.8
|
si:dkey-234l24.8 |
| chr15_+_5360407 | 0.22 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr15_-_14212777 | 0.03 |
ENSDART00000165572
|
CR925813.1
|
|
| chr24_+_17345521 | 0.01 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr21_-_13661631 | 0.01 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
Network of associatons between targets according to the STRING database.
First level regulatory network of msx1b+msx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.7 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 1.4 | 7.2 | GO:2000742 | regulation of anterior head development(GO:2000742) |
| 1.4 | 4.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 1.4 | 28.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 1.4 | 8.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.3 | 4.0 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 1.3 | 6.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.7 | 3.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.6 | 3.0 | GO:0070814 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.5 | 4.6 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.5 | 1.5 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.5 | 4.6 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.4 | 0.9 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.4 | 4.8 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.4 | 6.3 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.4 | 2.5 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.3 | 6.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.3 | 5.1 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.3 | 2.5 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 2.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 5.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.3 | 7.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.3 | 3.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 16.3 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.2 | 1.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.2 | 2.9 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 4.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 3.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 1.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 0.9 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.2 | 3.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 2.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 2.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 7.4 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 2.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 2.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 7.0 | GO:0072676 | lymphocyte migration(GO:0072676) |
| 0.1 | 1.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 7.0 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 2.1 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 2.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 3.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 3.4 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 10.4 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.1 | 3.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 2.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 9.9 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 4.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 5.5 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 1.2 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 1.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 3.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 9.0 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.0 | 4.0 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 2.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.3 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 2.6 | GO:0045165 | cell fate commitment(GO:0045165) |
| 0.0 | 1.0 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 5.6 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 3.4 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 2.0 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.1 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.8 | 5.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 2.7 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.4 | 2.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 2.0 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 26.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 1.7 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.2 | 1.9 | GO:0035060 | brahma complex(GO:0035060) |
| 0.2 | 4.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 4.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 4.7 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 2.0 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 4.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 3.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 13.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 4.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 3.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 4.0 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 6.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 2.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 5.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 15.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.5 | 6.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.3 | 28.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 1.1 | 3.3 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 1.0 | 4.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.6 | 3.0 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.5 | 2.7 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.5 | 10.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.5 | 7.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.5 | 7.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 5.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.4 | 4.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 2.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 7.0 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.2 | 5.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.9 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 2.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 3.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 4.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 3.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 3.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 2.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 5.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 2.0 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 4.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 4.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.6 | GO:0010181 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) FMN binding(GO:0010181) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 4.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 7.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 21.7 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 2.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 7.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.7 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 22.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 7.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 5.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 4.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 7.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 2.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 8.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 4.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 5.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 6.2 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 1.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 4.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |