Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
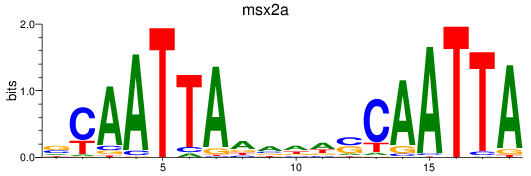
Results for msx2a
Z-value: 0.89
Transcription factors associated with msx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx2a
|
ENSDARG00000104651 | muscle segment homeobox 2a |
|
msx2a
|
ENSDARG00000115813 | muscle segment homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx2a | dr11_v1_chr14_-_24081929_24081929 | -0.61 | 6.7e-11 | Click! |
Activity profile of msx2a motif
Sorted Z-values of msx2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_53374454 | 20.73 |
ENSDART00000038807
|
ucp1
|
uncoupling protein 1 |
| chr5_+_2815021 | 19.70 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr1_+_10051763 | 17.15 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr24_-_38110779 | 10.21 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr21_+_5589923 | 8.72 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr20_+_38032143 | 7.03 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr20_+_38031439 | 6.95 |
ENSDART00000153208
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr5_-_20123002 | 6.92 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr1_+_29068654 | 6.85 |
ENSDART00000053932
|
cbsa
|
cystathionine-beta-synthase a |
| chr7_+_56577522 | 6.63 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr22_-_24818066 | 5.87 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr7_+_27603211 | 5.60 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr7_+_56577906 | 5.51 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr14_+_29941445 | 5.16 |
ENSDART00000181761
|
fam149a
|
family with sequence similarity 149 member A |
| chr13_+_8693410 | 4.89 |
ENSDART00000138448
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr14_-_4076480 | 4.88 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr15_+_32405959 | 4.27 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr23_-_39784368 | 4.25 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr23_-_44786844 | 4.11 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr8_-_38616712 | 3.54 |
ENSDART00000141827
|
si:ch211-198d23.1
|
si:ch211-198d23.1 |
| chr4_-_1147655 | 3.35 |
ENSDART00000171561
|
si:ch211-117i20.2
|
si:ch211-117i20.2 |
| chr15_+_32423801 | 3.28 |
ENSDART00000165591
|
si:dkey-285b23.3
|
si:dkey-285b23.3 |
| chr16_-_26255877 | 2.96 |
ENSDART00000146214
|
erfl1
|
Ets2 repressor factor like 1 |
| chr6_+_34870374 | 2.89 |
ENSDART00000149356
|
il23r
|
interleukin 23 receptor |
| chr9_+_28598577 | 2.69 |
ENSDART00000142623
ENSDART00000135947 |
si:ch73-7i4.1
|
si:ch73-7i4.1 |
| chr15_-_29348212 | 2.68 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr23_-_44574059 | 2.55 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr12_+_47663419 | 2.53 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr19_+_31585341 | 2.35 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr3_+_16771797 | 2.34 |
ENSDART00000147418
|
lrrc3cb
|
leucine rich repeat containing 3Cb |
| chr15_+_29727799 | 2.26 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr15_+_22394074 | 2.21 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr16_-_13613475 | 2.09 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr8_-_34762163 | 2.03 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr13_+_33373713 | 2.01 |
ENSDART00000158709
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr6_+_8315050 | 1.99 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr14_+_29941266 | 1.97 |
ENSDART00000112757
|
fam149a
|
family with sequence similarity 149 member A |
| chr1_+_19764995 | 1.91 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr17_+_30448452 | 1.88 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr17_-_20118145 | 1.87 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr5_-_40040488 | 1.85 |
ENSDART00000157755
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr2_-_1569250 | 1.78 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr10_-_40484531 | 1.78 |
ENSDART00000142464
ENSDART00000180318 |
taar20z
|
trace amine associated receptor 20z |
| chr10_-_40448736 | 1.77 |
ENSDART00000137644
ENSDART00000168190 |
taar20p
|
trace amine associated receptor 20p |
| chr25_+_19095231 | 1.75 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr25_+_37126921 | 1.73 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr4_-_25271455 | 1.66 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr17_+_23729717 | 1.62 |
ENSDART00000179026
|
zgc:91976
|
zgc:91976 |
| chr21_+_8439729 | 1.62 |
ENSDART00000182825
ENSDART00000055336 |
dennd1a
|
DENN/MADD domain containing 1A |
| chr2_-_3611960 | 1.60 |
ENSDART00000184579
|
pter
|
phosphotriesterase related |
| chr10_-_40524741 | 1.58 |
ENSDART00000030565
|
taar18a
|
trace amine associated receptor 18a |
| chr17_+_23730089 | 1.56 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr10_+_40756352 | 1.52 |
ENSDART00000156210
ENSDART00000144576 |
taar19f
|
trace amine associated receptor 19f |
| chr7_+_59169081 | 1.49 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr2_-_41124013 | 1.48 |
ENSDART00000134756
|
hs6st1a
|
heparan sulfate 6-O-sulfotransferase 1a |
| chr10_-_40397354 | 1.45 |
ENSDART00000150775
ENSDART00000156420 |
taar20o
|
trace amine associated receptor 20o |
| chr7_-_51773166 | 1.42 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr15_-_34214440 | 1.42 |
ENSDART00000167052
|
etv1
|
ets variant 1 |
| chr10_-_40528229 | 1.40 |
ENSDART00000146927
ENSDART00000148354 |
taar18g
|
trace amine associated receptor 18g |
| chr16_-_46578523 | 1.40 |
ENSDART00000131061
|
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr13_+_18321140 | 1.39 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr6_-_40446536 | 1.38 |
ENSDART00000153466
|
tatdn2
|
TatD DNase domain containing 2 |
| chr3_-_50954607 | 1.35 |
ENSDART00000163810
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr8_+_40644838 | 1.32 |
ENSDART00000169311
|
adra2b
|
adrenoceptor alpha 2B |
| chr12_-_6063328 | 1.31 |
ENSDART00000002583
|
aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr10_+_40781052 | 1.31 |
ENSDART00000191061
ENSDART00000134435 |
taar19c
|
trace amine associated receptor 19c |
| chr1_-_17735861 | 1.30 |
ENSDART00000018238
|
pdlim3a
|
PDZ and LIM domain 3a |
| chr6_+_25257728 | 1.30 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr16_+_10918252 | 1.29 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr24_+_17269849 | 1.29 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr13_+_38226205 | 1.29 |
ENSDART00000144093
ENSDART00000140179 |
si:dkeyp-4c7.3
|
si:dkeyp-4c7.3 |
| chr24_-_32582880 | 1.28 |
ENSDART00000186307
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr10_-_40514643 | 1.28 |
ENSDART00000140705
|
taar19k
|
trace amine associated receptor 19k |
| chr10_-_40402904 | 1.28 |
ENSDART00000145101
|
taar20t
|
trace amine associated receptor 20t |
| chr6_+_30430591 | 1.27 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr10_-_40490647 | 1.27 |
ENSDART00000143660
|
taar20x
|
trace amine associated receptor 20x |
| chr10_-_40360635 | 1.27 |
ENSDART00000131970
|
taar20g
|
trace amine associated receptor 20g |
| chr10_+_40690411 | 1.25 |
ENSDART00000146865
|
taar19m
|
trace amine associated receptor 19m |
| chr15_-_576135 | 1.24 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr3_+_27929505 | 1.23 |
ENSDART00000055528
|
TMEM114
|
transmembrane protein 114 |
| chr10_-_40416054 | 1.22 |
ENSDART00000137947
ENSDART00000143606 |
taar20j
|
trace amine associated receptor 20j |
| chr16_+_28578648 | 1.22 |
ENSDART00000149566
|
nmt2
|
N-myristoyltransferase 2 |
| chr19_-_11315224 | 1.19 |
ENSDART00000104933
|
eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr4_-_6459863 | 1.17 |
ENSDART00000138367
|
foxp2
|
forkhead box P2 |
| chr10_+_40774215 | 1.16 |
ENSDART00000131493
|
taar19b
|
trace amine associated receptor 19b |
| chr10_+_40762200 | 1.16 |
ENSDART00000161000
ENSDART00000136185 |
taar19g
|
trace amine associated receptor 19g |
| chr21_-_21089781 | 1.16 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr23_-_900795 | 1.16 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr10_+_40742685 | 1.14 |
ENSDART00000184858
ENSDART00000140300 |
taar19e
|
trace amine associated receptor 19e |
| chr24_+_11381400 | 1.12 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr19_-_10612774 | 1.11 |
ENSDART00000091898
|
si:dkey-211g8.6
|
si:dkey-211g8.6 |
| chr20_-_9436521 | 1.10 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr24_+_17270129 | 1.09 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr25_+_35891342 | 1.06 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr10_-_40406278 | 1.05 |
ENSDART00000131933
ENSDART00000156945 |
taar20i
|
trace amine associated receptor 20i |
| chr14_-_1998501 | 1.04 |
ENSDART00000189052
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr10_-_40508527 | 1.04 |
ENSDART00000121514
ENSDART00000133729 |
taar20b1
|
trace amine associated receptor 20b1 |
| chr8_+_41048501 | 1.02 |
ENSDART00000123288
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr2_-_10192459 | 1.02 |
ENSDART00000128535
ENSDART00000017173 |
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr19_-_30447611 | 1.02 |
ENSDART00000073705
ENSDART00000048977 ENSDART00000191237 |
abcf1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr9_-_7652792 | 1.02 |
ENSDART00000137957
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr7_+_4474880 | 1.02 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr10_-_40366360 | 1.02 |
ENSDART00000188830
ENSDART00000192043 ENSDART00000150415 |
taar20i
|
trace amine associated receptor 20i |
| chr24_-_7699356 | 1.01 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr10_+_40768203 | 1.01 |
ENSDART00000171994
ENSDART00000140343 |
taar19d
|
trace amine associated receptor 19d |
| chr10_-_20524387 | 1.01 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr10_+_40633990 | 1.00 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr10_-_40454685 | 0.99 |
ENSDART00000137832
|
taar20o
|
trace amine associated receptor 20o |
| chr10_+_40731834 | 0.98 |
ENSDART00000182900
ENSDART00000139284 |
taar19i
|
trace amine associated receptor 19i |
| chr17_+_35362851 | 0.98 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr10_+_40711286 | 0.98 |
ENSDART00000140242
|
si:ch211-139n6.11
|
si:ch211-139n6.11 |
| chr2_+_38608290 | 0.97 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr4_+_19700308 | 0.95 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr7_+_23515966 | 0.94 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr10_-_34870667 | 0.94 |
ENSDART00000161272
|
dclk1a
|
doublecortin-like kinase 1a |
| chr17_-_31483469 | 0.93 |
ENSDART00000062907
ENSDART00000061547 |
ltk
|
leukocyte receptor tyrosine kinase |
| chr10_+_40737540 | 0.92 |
ENSDART00000125577
|
taar19a
|
trace amine associated receptor 19a |
| chr19_-_868187 | 0.91 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr2_-_54039293 | 0.91 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr10_+_40660772 | 0.90 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr13_-_37109987 | 0.89 |
ENSDART00000136750
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr10_+_40665182 | 0.89 |
ENSDART00000099156
|
taar19u
|
trace amine associated receptor 19u |
| chr10_-_40443722 | 0.88 |
ENSDART00000132906
ENSDART00000157598 |
taar20q
|
trace amine associated receptor 20q |
| chr7_-_49351706 | 0.84 |
ENSDART00000174151
|
brsk2b
|
BR serine/threonine kinase 2b |
| chr10_-_20524592 | 0.84 |
ENSDART00000185048
|
ddhd2
|
DDHD domain containing 2 |
| chr15_-_6966221 | 0.83 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr5_-_18962794 | 0.82 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr6_+_18359306 | 0.81 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr1_+_8442071 | 0.81 |
ENSDART00000143547
|
myo15ab
|
myosin XVAb |
| chr10_-_40479911 | 0.81 |
ENSDART00000136741
|
taar20d1
|
trace amine associated receptor 20d1 |
| chr10_-_29900546 | 0.80 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr24_-_20926812 | 0.80 |
ENSDART00000130958
|
ccdc58
|
coiled-coil domain containing 58 |
| chr10_+_40722256 | 0.79 |
ENSDART00000099154
|
taar19q
|
trace amine associated receptor 19q |
| chr16_+_51326237 | 0.79 |
ENSDART00000168525
|
CU469384.1
|
|
| chr7_+_13918349 | 0.78 |
ENSDART00000172772
ENSDART00000173384 |
si:cabz01059983.1
|
si:cabz01059983.1 |
| chr10_+_40726399 | 0.78 |
ENSDART00000142812
|
taar19j
|
trace amine associated receptor 19j |
| chr11_-_12471837 | 0.78 |
ENSDART00000113175
ENSDART00000186965 |
si:dkey-27d5.6
|
si:dkey-27d5.6 |
| chr21_-_929293 | 0.77 |
ENSDART00000006419
|
txnl1
|
thioredoxin-like 1 |
| chr10_-_40333319 | 0.75 |
ENSDART00000150479
|
taar20a
|
trace amine associated receptor 20a |
| chr24_+_40473670 | 0.75 |
ENSDART00000180163
|
CABZ01076968.1
|
|
| chr10_+_40629616 | 0.74 |
ENSDART00000147476
|
CR396590.9
|
|
| chr17_+_1496107 | 0.72 |
ENSDART00000187804
|
LO018430.1
|
|
| chr10_-_42751641 | 0.72 |
ENSDART00000182734
ENSDART00000113926 |
zgc:100918
|
zgc:100918 |
| chr14_-_22385779 | 0.71 |
ENSDART00000192211
ENSDART00000175309 |
fbxl3l
|
F-box and leucine-rich repeat protein 3, like |
| chr17_+_5768608 | 0.71 |
ENSDART00000157039
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr10_-_40503395 | 0.70 |
ENSDART00000134499
ENSDART00000193101 |
si:dkey-181m19.17
|
si:dkey-181m19.17 |
| chr5_+_22177033 | 0.70 |
ENSDART00000131223
|
fdx1b
|
ferredoxin 1b |
| chr11_-_12472006 | 0.70 |
ENSDART00000160558
|
si:dkey-27d5.6
|
si:dkey-27d5.6 |
| chr11_-_165288 | 0.70 |
ENSDART00000108703
ENSDART00000173151 |
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
| chr1_+_26480890 | 0.70 |
ENSDART00000164430
|
uso1
|
USO1 vesicle transport factor |
| chr15_-_28094256 | 0.68 |
ENSDART00000142041
ENSDART00000132153 ENSDART00000146657 ENSDART00000048720 |
cryba1a
|
crystallin, beta A1a |
| chr3_-_58226583 | 0.66 |
ENSDART00000187429
|
im:6904045
|
im:6904045 |
| chr6_-_16717878 | 0.66 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr25_+_14507567 | 0.66 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr22_+_12366516 | 0.66 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr13_+_18331509 | 0.64 |
ENSDART00000181918
|
zgc:110319
|
zgc:110319 |
| chr16_-_13921589 | 0.63 |
ENSDART00000023543
|
rcvrn2
|
recoverin 2 |
| chr10_-_40543896 | 0.63 |
ENSDART00000186921
ENSDART00000136031 |
taar18f
|
trace amine associated receptor 18f |
| chr10_-_40474628 | 0.62 |
ENSDART00000133892
|
taar20a1
|
trace amine associated receptor 20a1 |
| chr23_+_44157682 | 0.62 |
ENSDART00000164474
ENSDART00000149928 |
si:ch73-106g13.1
|
si:ch73-106g13.1 |
| chr10_-_40429747 | 0.60 |
ENSDART00000150613
|
taar20r
|
trace amine associated receptor 20r |
| chr9_+_53337974 | 0.60 |
ENSDART00000145138
|
dct
|
dopachrome tautomerase |
| chr22_-_9183944 | 0.60 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr14_-_43616572 | 0.60 |
ENSDART00000111189
|
gar1
|
GAR1 homolog, ribonucleoprotein |
| chr20_+_22220988 | 0.60 |
ENSDART00000049204
|
kdr
|
kinase insert domain receptor (a type III receptor tyrosine kinase) |
| chr10_+_40700311 | 0.60 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr19_+_43604643 | 0.59 |
ENSDART00000151168
|
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr21_-_929448 | 0.58 |
ENSDART00000133976
|
txnl1
|
thioredoxin-like 1 |
| chr17_+_24613255 | 0.58 |
ENSDART00000064738
|
atp5if1b
|
ATP synthase inhibitory factor subunit 1b |
| chr4_+_5796761 | 0.57 |
ENSDART00000164854
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr11_+_17984167 | 0.57 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr14_+_45675306 | 0.56 |
ENSDART00000105461
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr10_-_5847655 | 0.56 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr16_+_34111919 | 0.55 |
ENSDART00000134037
ENSDART00000006061 ENSDART00000140552 |
tcea3
|
transcription elongation factor A (SII), 3 |
| chr8_+_10561922 | 0.55 |
ENSDART00000133348
|
fam19a5l
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5-like |
| chr10_-_42108137 | 0.55 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr19_-_31584444 | 0.55 |
ENSDART00000052183
|
zgc:111986
|
zgc:111986 |
| chr21_-_16113477 | 0.55 |
ENSDART00000147588
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr16_-_41646164 | 0.54 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr17_-_36841981 | 0.54 |
ENSDART00000131566
|
myo6b
|
myosin VIb |
| chr10_-_5847904 | 0.53 |
ENSDART00000161096
|
ankrd55
|
ankyrin repeat domain 55 |
| chr17_+_43032529 | 0.51 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr6_-_39270851 | 0.51 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr4_+_2267641 | 0.51 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr20_+_26556372 | 0.50 |
ENSDART00000187179
ENSDART00000157291 |
irf4b
|
interferon regulatory factor 4b |
| chr22_+_1313046 | 0.49 |
ENSDART00000170421
|
si:ch73-138e16.6
|
si:ch73-138e16.6 |
| chr9_-_55790458 | 0.48 |
ENSDART00000161247
|
sowahca
|
sosondowah ankyrin repeat domain family Ca |
| chr12_-_19007834 | 0.47 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr8_+_9866351 | 0.46 |
ENSDART00000133985
|
kcnd1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr5_+_56268436 | 0.44 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr10_+_19310688 | 0.44 |
ENSDART00000141364
|
lrrtm4l1
|
leucine rich repeat transmembrane neuronal 4 like 1 |
| chr12_-_29305533 | 0.44 |
ENSDART00000189410
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr19_+_43604256 | 0.44 |
ENSDART00000151080
ENSDART00000110305 |
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr9_-_32753535 | 0.43 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr11_-_2632747 | 0.42 |
ENSDART00000008133
|
si:ch211-160o17.2
|
si:ch211-160o17.2 |
| chr22_+_26853254 | 0.41 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr23_-_1660708 | 0.41 |
ENSDART00000175138
|
CU693481.1
|
|
| chr6_+_55174744 | 0.41 |
ENSDART00000023562
|
SYT2
|
synaptotagmin 2 |
| chr14_-_1987121 | 0.40 |
ENSDART00000135733
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr25_+_10909850 | 0.40 |
ENSDART00000186021
|
CR339041.3
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of msx2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 19.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 2.6 | 20.7 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 1.6 | 4.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 1.4 | 6.9 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 1.1 | 17.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.9 | 5.6 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.8 | 2.3 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.7 | 2.7 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.5 | 4.8 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.4 | 3.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.4 | 1.4 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.3 | 1.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.3 | 1.0 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.3 | 0.9 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 1.3 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.2 | 2.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 1.8 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 1.9 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 0.7 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.2 | 1.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 5.9 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.2 | 3.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 1.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 1.4 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 1.4 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.7 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.9 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 1.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.5 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.4 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.7 | GO:0034650 | glucocorticoid biosynthetic process(GO:0006704) cortisol metabolic process(GO:0034650) |
| 0.1 | 1.3 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.1 | 0.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 2.0 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 | 2.0 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.2 | GO:0051574 | positive regulation of histone methylation(GO:0031062) positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.8 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 1.9 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.6 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.0 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 1.0 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 1.9 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 0.3 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 2.0 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 15.5 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 3.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.4 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.4 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 27.3 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 1.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.6 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.1 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 2.8 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.3 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 2.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.6 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 2.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.3 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 36.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 17.6 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 3.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 3.5 | GO:0030425 | dendrite(GO:0030425) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.7 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 5.2 | 20.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.7 | 8.7 | GO:2001070 | starch binding(GO:2001070) |
| 1.4 | 6.9 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.8 | 5.9 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.8 | 2.3 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.5 | 3.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.5 | 2.0 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.5 | 33.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.5 | 1.9 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.4 | 1.9 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 1.4 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.3 | 1.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.3 | 2.0 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.2 | 1.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 1.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 1.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 10.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 3.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 5.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.5 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 1.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.4 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 4.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 1.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.4 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 14.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 11.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 3.4 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 2.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.9 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 17.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 2.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.3 | 10.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 17.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.9 | 10.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 14.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.3 | 19.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.2 | 5.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |