Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
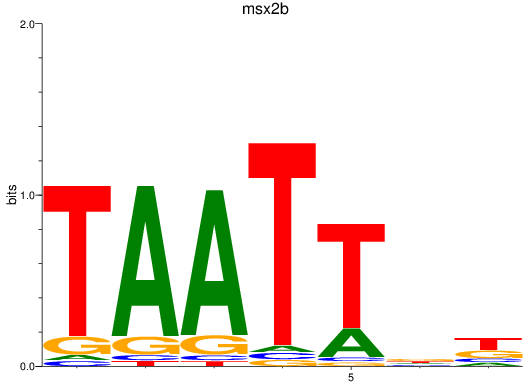
Results for msx2b
Z-value: 0.61
Transcription factors associated with msx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx2b
|
ENSDARG00000101023 | muscle segment homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx2b | dr11_v1_chr21_-_41147818_41147818 | -0.22 | 3.1e-02 | Click! |
Activity profile of msx2b motif
Sorted Z-values of msx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_37178271 | 6.11 |
ENSDART00000161771
|
itih3b
|
inter-alpha-trypsin inhibitor heavy chain 3b |
| chr13_+_23988442 | 5.27 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr13_+_8958438 | 4.59 |
ENSDART00000165721
ENSDART00000169398 |
haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr10_+_25726694 | 3.91 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr14_+_1007169 | 3.63 |
ENSDART00000172676
|
f8
|
coagulation factor VIII, procoagulant component |
| chr21_+_38002879 | 3.35 |
ENSDART00000065183
|
cldn2
|
claudin 2 |
| chr8_+_40477264 | 3.20 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr15_-_34408777 | 3.05 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr4_+_14900042 | 2.66 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr14_-_11456724 | 2.55 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr18_+_7073130 | 2.17 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr20_+_38031439 | 2.17 |
ENSDART00000153208
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr24_+_26017094 | 2.15 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr5_+_24179307 | 1.98 |
ENSDART00000051552
|
mpdu1a
|
mannose-P-dolichol utilization defect 1a |
| chr13_+_23132666 | 1.98 |
ENSDART00000164639
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr10_-_25217347 | 1.89 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr8_+_20140321 | 1.84 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr9_-_24031461 | 1.83 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr10_+_6013076 | 1.71 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr20_+_36628059 | 1.63 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr5_-_62317496 | 1.62 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr13_-_4223955 | 1.60 |
ENSDART00000113060
|
dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr7_+_6990510 | 1.60 |
ENSDART00000138948
|
ccs
|
copper chaperone for superoxide dismutase |
| chr18_-_26715655 | 1.59 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr5_-_38777852 | 1.51 |
ENSDART00000131603
|
si:dkey-58f10.4
|
si:dkey-58f10.4 |
| chr15_+_17031111 | 1.50 |
ENSDART00000175378
|
plin2
|
perilipin 2 |
| chr12_-_42368296 | 1.49 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr21_-_35082715 | 1.46 |
ENSDART00000146454
|
adrb2b
|
adrenoceptor beta 2, surface b |
| chr2_-_24398324 | 1.43 |
ENSDART00000165226
|
zgc:154006
|
zgc:154006 |
| chr7_-_61901695 | 1.32 |
ENSDART00000110328
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr21_+_10712823 | 1.26 |
ENSDART00000123476
|
lman1
|
lectin, mannose-binding, 1 |
| chr15_+_17074185 | 1.23 |
ENSDART00000046648
|
clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr19_-_22346582 | 1.21 |
ENSDART00000045675
ENSDART00000169065 |
slc52a2
zgc:109744
|
solute carrier family 52 (riboflavin transporter), member 2 zgc:109744 |
| chr20_-_23426339 | 1.20 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr17_+_8542203 | 1.17 |
ENSDART00000158873
|
CU462878.2
|
|
| chr5_-_9216758 | 1.17 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr15_+_7064819 | 1.12 |
ENSDART00000155268
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr7_-_59210882 | 1.08 |
ENSDART00000170330
ENSDART00000158996 |
nagk
|
N-acetylglucosamine kinase |
| chr16_+_41570653 | 1.08 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr2_-_50225411 | 1.07 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr13_+_28747934 | 1.07 |
ENSDART00000160176
ENSDART00000101633 ENSDART00000136318 |
prom2
|
prominin 2 |
| chr1_+_43686251 | 1.04 |
ENSDART00000074604
ENSDART00000137791 |
cisd2
|
CDGSH iron sulfur domain 2 |
| chr15_+_17030941 | 1.03 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr2_+_42871831 | 1.03 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr2_-_10600950 | 1.00 |
ENSDART00000160216
|
BX323564.1
|
|
| chr16_-_50897887 | 1.00 |
ENSDART00000156985
|
si:ch73-90p23.1
|
si:ch73-90p23.1 |
| chr9_+_41156818 | 0.98 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr15_-_19051152 | 0.98 |
ENSDART00000186453
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr17_-_18898115 | 0.97 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr6_+_54239625 | 0.93 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr11_-_44484952 | 0.91 |
ENSDART00000166674
ENSDART00000188016 |
mfn1b
|
mitofusin 1b |
| chr25_+_30442389 | 0.85 |
ENSDART00000003346
|
pdcd2l
|
programmed cell death 2-like |
| chr14_+_35369979 | 0.85 |
ENSDART00000144702
ENSDART00000169712 |
clint1a
|
clathrin interactor 1a |
| chr22_-_16755885 | 0.85 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr12_-_3992839 | 0.84 |
ENSDART00000126100
|
si:ch211-136m16.8
|
si:ch211-136m16.8 |
| chr18_-_16907436 | 0.83 |
ENSDART00000146464
|
swap70b
|
switching B cell complex subunit SWAP70b |
| chr3_-_13509283 | 0.83 |
ENSDART00000166832
|
slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr14_-_8453192 | 0.81 |
ENSDART00000136947
|
eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr23_-_44903048 | 0.79 |
ENSDART00000149103
|
fhdc5
|
FH2 domain containing 5 |
| chr19_-_7441948 | 0.79 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr19_-_12392819 | 0.78 |
ENSDART00000052239
|
ptpn2b
|
protein tyrosine phosphatase, non-receptor type 2, b |
| chr11_+_17984167 | 0.77 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr4_-_5455506 | 0.77 |
ENSDART00000156593
ENSDART00000154676 |
si:dkey-14d8.22
|
si:dkey-14d8.22 |
| chr6_+_8630355 | 0.75 |
ENSDART00000161749
ENSDART00000193976 |
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr19_+_12406583 | 0.75 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr17_+_21477892 | 0.72 |
ENSDART00000155309
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr6_+_8626427 | 0.71 |
ENSDART00000193660
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr23_-_14990865 | 0.69 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr7_+_33145529 | 0.68 |
ENSDART00000052387
|
itln2
|
intelectin 2 |
| chr25_-_28926631 | 0.67 |
ENSDART00000112850
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr11_-_6880725 | 0.66 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr22_-_18164671 | 0.66 |
ENSDART00000014057
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr11_+_17984354 | 0.66 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr2_-_26590628 | 0.66 |
ENSDART00000025120
|
ndc1
|
NDC1 transmembrane nucleoporin |
| chr8_+_23738122 | 0.63 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr22_-_18164835 | 0.63 |
ENSDART00000143189
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr3_+_22935183 | 0.60 |
ENSDART00000157378
|
hdac5
|
histone deacetylase 5 |
| chr3_-_44059902 | 0.60 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr1_-_24462544 | 0.58 |
ENSDART00000184421
ENSDART00000184987 ENSDART00000126950 |
sh3d19
|
SH3 domain containing 19 |
| chr4_+_12966640 | 0.58 |
ENSDART00000113357
|
vhll
|
von Hippel-Lindau tumor suppressor like |
| chr6_+_6828167 | 0.58 |
ENSDART00000181284
|
si:ch211-85n16.4
|
si:ch211-85n16.4 |
| chr10_-_16065185 | 0.56 |
ENSDART00000187266
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr7_-_64971839 | 0.55 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr17_+_1496107 | 0.49 |
ENSDART00000187804
|
LO018430.1
|
|
| chr22_-_15717897 | 0.49 |
ENSDART00000008424
|
malt2
|
MALT paracaspase 2 |
| chr13_-_36050303 | 0.49 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr7_+_15313443 | 0.47 |
ENSDART00000045385
|
mespba
|
mesoderm posterior ba |
| chr15_-_17813680 | 0.46 |
ENSDART00000158556
|
CT573342.2
|
|
| chr1_-_45213565 | 0.46 |
ENSDART00000145757
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr4_+_17671164 | 0.45 |
ENSDART00000040445
|
gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr2_+_37176814 | 0.45 |
ENSDART00000048277
ENSDART00000176827 |
copa
|
coatomer protein complex, subunit alpha |
| chr11_+_25539698 | 0.45 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr8_-_11324143 | 0.44 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr21_-_5205617 | 0.44 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr13_-_25819825 | 0.43 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr20_+_35484070 | 0.41 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr3_-_33427803 | 0.41 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr22_+_1887750 | 0.40 |
ENSDART00000180917
|
si:dkey-15h8.17
|
si:dkey-15h8.17 |
| chr12_+_10115964 | 0.40 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr8_-_1838315 | 0.40 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr6_-_45869127 | 0.40 |
ENSDART00000062459
ENSDART00000180563 |
rbm19
|
RNA binding motif protein 19 |
| chr25_+_10909850 | 0.38 |
ENSDART00000186021
|
CR339041.3
|
|
| chr5_-_61609448 | 0.38 |
ENSDART00000133426
|
si:dkey-261j4.5
|
si:dkey-261j4.5 |
| chr21_+_4313039 | 0.37 |
ENSDART00000141146
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr13_+_50151407 | 0.37 |
ENSDART00000031858
|
gpr137ba
|
G protein-coupled receptor 137Ba |
| chr13_+_31583034 | 0.34 |
ENSDART00000111763
|
six6a
|
SIX homeobox 6a |
| chr14_+_23874062 | 0.33 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr12_+_46404307 | 0.32 |
ENSDART00000185011
|
BX005305.4
|
|
| chr5_+_24882633 | 0.32 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr11_+_44804685 | 0.32 |
ENSDART00000163660
|
strn
|
striatin, calmodulin binding protein |
| chr9_+_29548195 | 0.31 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr22_-_30973791 | 0.31 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr19_-_1947403 | 0.30 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr15_-_23784600 | 0.29 |
ENSDART00000059354
|
rad1
|
RAD1 homolog (S. pombe) |
| chr5_-_66768121 | 0.29 |
ENSDART00000141095
|
FERMT3 (1 of many)
|
im:7154036 |
| chr1_+_54137089 | 0.29 |
ENSDART00000062945
|
LO017798.1
|
|
| chr22_-_23228669 | 0.28 |
ENSDART00000079536
|
nek7
|
NIMA-related kinase 7 |
| chr13_+_2625150 | 0.27 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr19_+_24891747 | 0.26 |
ENSDART00000132209
ENSDART00000193610 |
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr5_-_4297459 | 0.26 |
ENSDART00000018895
|
srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr19_-_3255725 | 0.26 |
ENSDART00000105165
|
illr1
|
immune-related, lectin-like receptor 1 |
| chr13_-_40238813 | 0.26 |
ENSDART00000044963
|
loxl4
|
lysyl oxidase-like 4 |
| chr11_+_2687395 | 0.26 |
ENSDART00000082510
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr14_-_899170 | 0.26 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr21_+_27513859 | 0.26 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr10_-_11385155 | 0.25 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr23_-_31913231 | 0.25 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr23_-_24493768 | 0.25 |
ENSDART00000135559
ENSDART00000046933 |
sult1st5
|
sulfotransferase family 1, cytosolic sulfotransferase 5 |
| chr3_-_31619463 | 0.24 |
ENSDART00000124559
|
moto
|
minamoto |
| chr17_-_21162821 | 0.23 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr14_-_3132188 | 0.22 |
ENSDART00000172259
|
si:zfos-1069f5.1
|
si:zfos-1069f5.1 |
| chr17_-_200316 | 0.22 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr23_-_32092443 | 0.22 |
ENSDART00000133688
|
letmd1
|
LETM1 domain containing 1 |
| chr10_+_16165533 | 0.20 |
ENSDART00000065045
|
prrc1
|
proline-rich coiled-coil 1 |
| chr3_-_34098731 | 0.20 |
ENSDART00000150999
|
ighv5-3
|
immunoglobulin heavy variable 5-3 |
| chr18_-_33979422 | 0.19 |
ENSDART00000136535
ENSDART00000167698 |
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr24_-_3783497 | 0.19 |
ENSDART00000158354
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr24_-_40860603 | 0.19 |
ENSDART00000188032
|
CU633479.7
|
|
| chr3_-_13461361 | 0.18 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr1_+_29183962 | 0.18 |
ENSDART00000113735
|
cars2
|
cysteinyl-tRNA synthetase 2, mitochondrial |
| chr5_+_66433287 | 0.18 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr15_-_28082310 | 0.18 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr3_-_17871846 | 0.18 |
ENSDART00000074478
ENSDART00000187941 |
nkiras2
|
NFKB inhibitor interacting Ras-like 2 |
| chr7_-_24005268 | 0.18 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr23_-_39772067 | 0.18 |
ENSDART00000159519
ENSDART00000189046 |
im:6912630
|
im:6912630 |
| chr25_-_24248000 | 0.18 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr23_+_43489182 | 0.17 |
ENSDART00000162062
|
znf341
|
zinc finger protein 341 |
| chr10_+_40660772 | 0.17 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr18_+_33264609 | 0.17 |
ENSDART00000050639
|
v2ra20
|
vomeronasal 2 receptor, a20 |
| chr15_-_4094888 | 0.16 |
ENSDART00000166307
|
TM4SF19
|
si:dkey-83h2.3 |
| chr5_-_29512538 | 0.16 |
ENSDART00000098364
|
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr10_-_5016997 | 0.15 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr23_-_41965557 | 0.14 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr11_+_25259058 | 0.13 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr17_-_25649079 | 0.12 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr6_-_27891961 | 0.11 |
ENSDART00000155116
|
im:7152348
|
im:7152348 |
| chr3_+_13848226 | 0.11 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr18_+_10884996 | 0.11 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr17_-_8656155 | 0.10 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr23_-_31913069 | 0.09 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr17_+_5915875 | 0.09 |
ENSDART00000184179
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr18_-_45761868 | 0.09 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr17_-_50054025 | 0.08 |
ENSDART00000190105
|
FO834825.3
|
|
| chr4_+_5506952 | 0.08 |
ENSDART00000032857
ENSDART00000160222 |
mapk11
|
mitogen-activated protein kinase 11 |
| chr14_-_7137808 | 0.08 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr8_+_37527575 | 0.08 |
ENSDART00000147239
|
or135-1
|
odorant receptor, family H, subfamily 135, member 1 |
| chr14_+_23520986 | 0.08 |
ENSDART00000170473
ENSDART00000175970 |
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr5_+_9246458 | 0.08 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr15_+_5339056 | 0.08 |
ENSDART00000174319
|
CABZ01018874.2
|
|
| chr10_+_5954787 | 0.07 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr25_+_25085349 | 0.07 |
ENSDART00000192166
|
si:ch73-182e20.4
|
si:ch73-182e20.4 |
| chr21_-_31013817 | 0.06 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr21_+_19062124 | 0.06 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr6_-_15641686 | 0.05 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr18_-_33979693 | 0.05 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr14_-_32089117 | 0.05 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr12_-_2993095 | 0.05 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr5_-_2689753 | 0.04 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr25_-_27729046 | 0.04 |
ENSDART00000131437
|
zgc:153935
|
zgc:153935 |
| chr22_+_31295791 | 0.03 |
ENSDART00000092447
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr13_-_50247689 | 0.03 |
ENSDART00000034541
|
gpatch11
|
G patch domain containing 11 |
| chr7_-_26532089 | 0.03 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr15_+_2190229 | 0.02 |
ENSDART00000147710
|
rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr10_-_41157135 | 0.02 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr17_-_10122204 | 0.02 |
ENSDART00000160751
|
BX088587.1
|
|
| chr10_-_18463934 | 0.02 |
ENSDART00000133116
ENSDART00000113422 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr2_-_23778180 | 0.01 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr16_-_35394422 | 0.01 |
ENSDART00000180593
|
si:dkey-34d22.3
|
si:dkey-34d22.3 |
| chr20_-_2134620 | 0.01 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr6_-_52348562 | 0.01 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr1_+_11977426 | 0.01 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
Network of associatons between targets according to the STRING database.
First level regulatory network of msx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.6 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.6 | 3.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.6 | 1.8 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.6 | 1.7 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.5 | 2.2 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.5 | 1.6 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.3 | 3.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.3 | 5.3 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.3 | 1.5 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.3 | 2.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 0.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.3 | 1.0 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.3 | 2.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 2.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 1.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 0.7 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.2 | 1.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 0.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.6 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 1.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.6 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.1 | 3.6 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 0.8 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.5 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 1.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.9 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 1.1 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 4.8 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 1.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.5 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 1.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.9 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.1 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.4 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0021795 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.0 | 0.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.3 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 1.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.0 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.6 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.5 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.9 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 2.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 5.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.9 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.5 | 1.6 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.5 | 2.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.5 | 3.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 1.5 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.3 | 3.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 1.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 1.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 1.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.8 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 4.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.9 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.8 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 1.1 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 1.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 3.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 3.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.6 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 2.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 5.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 1.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 2.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 5.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 4.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 3.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 3.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 9.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.1 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |