Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
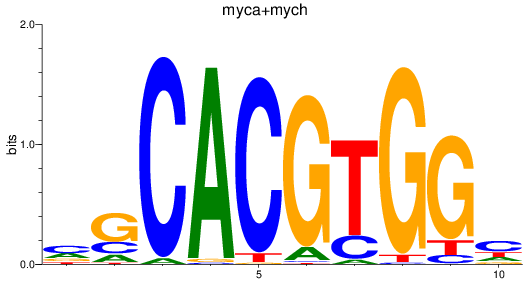
Results for myca+mych
Z-value: 0.77
Transcription factors associated with myca+mych
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myca
|
ENSDARG00000045695 | MYC proto-oncogene, bHLH transcription factor a |
|
mych
|
ENSDARG00000077473 | myelocytomatosis oncogene homolog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myca | dr11_v1_chr24_+_10413484_10413484 | -0.47 | 1.6e-06 | Click! |
| mych | dr11_v1_chr6_+_50451337_50451337 | 0.18 | 7.5e-02 | Click! |
Activity profile of myca+mych motif
Sorted Z-values of myca+mych motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_39764995 | 9.54 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr21_+_11969603 | 6.67 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr20_-_53996193 | 6.61 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr12_-_33359654 | 6.32 |
ENSDART00000001907
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr18_+_5490668 | 6.22 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr23_-_31555696 | 6.16 |
ENSDART00000053539
|
tcf21
|
transcription factor 21 |
| chr14_+_22076596 | 5.86 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr12_-_33359052 | 5.71 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr7_-_73752955 | 5.63 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr6_-_21189295 | 4.88 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr12_+_30586599 | 4.73 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr19_+_42983613 | 4.54 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr18_+_8917766 | 4.54 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr14_+_23518110 | 4.49 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr21_-_20765338 | 4.48 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr5_-_3927692 | 4.45 |
ENSDART00000146840
ENSDART00000058346 |
c1qbp
|
complement component 1, q subcomponent binding protein |
| chr7_+_67467702 | 4.35 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr12_-_33357655 | 4.29 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr22_-_26834043 | 4.15 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr8_+_44783424 | 4.15 |
ENSDART00000025875
|
si:ch1073-459j12.1
|
si:ch1073-459j12.1 |
| chr16_+_25245857 | 4.06 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr14_+_94603 | 4.05 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr13_-_12021566 | 4.05 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr19_+_43119698 | 3.83 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr16_+_16969060 | 3.70 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr16_+_16968682 | 3.66 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr18_-_11729 | 3.62 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr9_-_12888082 | 3.58 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr7_+_54605640 | 3.28 |
ENSDART00000168474
|
fgf3
|
fibroblast growth factor 3 |
| chr3_-_18805225 | 3.27 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr19_-_34117056 | 3.27 |
ENSDART00000158677
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr16_-_38629208 | 3.15 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr3_+_24595922 | 2.98 |
ENSDART00000169405
|
si:dkey-68o6.5
|
si:dkey-68o6.5 |
| chr21_+_26071874 | 2.98 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr13_-_3516473 | 2.96 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr20_+_54079341 | 2.85 |
ENSDART00000060444
|
rps29
|
ribosomal protein S29 |
| chr6_-_41536323 | 2.80 |
ENSDART00000113157
|
hemk1
|
HemK methyltransferase family member 1 |
| chr10_-_39130839 | 2.76 |
ENSDART00000061274
ENSDART00000148648 |
rps25
|
ribosomal protein S25 |
| chr24_+_34069675 | 2.75 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr22_-_607812 | 2.69 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr4_-_2945306 | 2.67 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr7_-_23745984 | 2.65 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr23_+_32499916 | 2.55 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr2_-_6182098 | 2.52 |
ENSDART00000156167
|
si:ch73-182a11.2
|
si:ch73-182a11.2 |
| chr22_-_5323482 | 2.51 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr5_-_69314495 | 2.50 |
ENSDART00000182335
|
smtnb
|
smoothelin b |
| chr20_+_25626479 | 2.47 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr19_-_46957968 | 2.47 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr11_-_37995501 | 2.46 |
ENSDART00000192096
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr23_+_43870886 | 2.46 |
ENSDART00000102658
ENSDART00000149088 |
nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr17_-_29771639 | 2.45 |
ENSDART00000086201
|
ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr18_-_11595567 | 2.44 |
ENSDART00000098565
|
CRACR2A
|
calcium release activated channel regulator 2A |
| chr7_+_20344032 | 2.35 |
ENSDART00000144948
ENSDART00000138786 |
ponzr1
|
plac8 onzin related protein 1 |
| chr18_+_14277003 | 2.31 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr12_+_13091842 | 2.27 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr19_-_24555935 | 2.24 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr19_-_24555623 | 2.23 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr9_-_22281854 | 2.15 |
ENSDART00000146319
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr2_-_42375275 | 2.14 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr17_+_8212477 | 2.13 |
ENSDART00000064665
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr3_-_37148594 | 2.10 |
ENSDART00000140855
|
mlx
|
MLX, MAX dimerization protein |
| chr20_+_46660339 | 2.10 |
ENSDART00000016530
|
adcy3b
|
adenylate cyclase 3b |
| chr14_+_94946 | 2.05 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr6_-_52348562 | 2.04 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr16_-_42186093 | 2.03 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr2_-_24369087 | 2.03 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr5_+_3927989 | 2.02 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr12_-_30359031 | 2.00 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr9_-_27396404 | 1.99 |
ENSDART00000136412
ENSDART00000101401 |
tex30
|
testis expressed 30 |
| chr22_+_5120033 | 1.95 |
ENSDART00000169200
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr9_+_56422311 | 1.94 |
ENSDART00000171958
|
gpr39
|
G protein-coupled receptor 39 |
| chr13_-_31370184 | 1.94 |
ENSDART00000034829
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr21_+_6114709 | 1.93 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr8_+_26059677 | 1.93 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr21_-_35853245 | 1.86 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr20_+_37294112 | 1.82 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr15_+_6661343 | 1.81 |
ENSDART00000160136
|
nop53
|
NOP53 ribosome biogenesis factor |
| chr14_+_16151636 | 1.78 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr7_+_20030888 | 1.76 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr24_+_16905188 | 1.73 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr15_-_17099560 | 1.72 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr7_-_58843193 | 1.71 |
ENSDART00000167231
|
mrpl15
|
mitochondrial ribosomal protein L15 |
| chr13_-_35907768 | 1.70 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr21_-_30082414 | 1.68 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr17_+_2162916 | 1.67 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr5_-_32338866 | 1.66 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr12_+_48395693 | 1.65 |
ENSDART00000180362
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr20_-_54259780 | 1.62 |
ENSDART00000172631
|
fkbp3
|
FK506 binding protein 3 |
| chr9_+_56422517 | 1.61 |
ENSDART00000168620
|
gpr39
|
G protein-coupled receptor 39 |
| chr2_+_49860722 | 1.60 |
ENSDART00000144060
|
rpl37
|
ribosomal protein L37 |
| chr3_+_1150348 | 1.58 |
ENSDART00000148524
|
nol12
|
nucleolar protein 12 |
| chr10_+_22034477 | 1.57 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr13_-_35892051 | 1.50 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr6_-_58975010 | 1.50 |
ENSDART00000144911
ENSDART00000144514 |
mars
|
methionyl-tRNA synthetase |
| chr17_-_45370200 | 1.48 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr24_-_42090635 | 1.43 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr18_-_20608025 | 1.43 |
ENSDART00000090156
ENSDART00000151980 |
bcl2l13
|
BCL2 like 13 |
| chr13_+_31402067 | 1.41 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr22_+_17261801 | 1.38 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr21_+_34119759 | 1.38 |
ENSDART00000024750
ENSDART00000128242 |
hmgb3b
|
high mobility group box 3b |
| chr6_-_52796212 | 1.37 |
ENSDART00000154133
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr23_-_38160024 | 1.37 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr1_-_55750208 | 1.35 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr15_-_36533322 | 1.35 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr11_+_6009984 | 1.32 |
ENSDART00000185680
|
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr9_-_30259295 | 1.32 |
ENSDART00000139106
|
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr16_+_40954481 | 1.29 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr22_-_30430276 | 1.29 |
ENSDART00000123840
|
BX649448.1
|
|
| chr20_+_25445826 | 1.28 |
ENSDART00000012581
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr5_-_16425781 | 1.28 |
ENSDART00000185624
ENSDART00000180617 |
slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr9_+_38158570 | 1.27 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr7_+_30779761 | 1.26 |
ENSDART00000066806
ENSDART00000173671 |
mcee
|
methylmalonyl CoA epimerase |
| chr18_+_33386372 | 1.26 |
ENSDART00000141838
|
v2rh12
|
vomeronasal 2 receptor, h12 |
| chr13_+_30912385 | 1.26 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr14_+_5385855 | 1.24 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr20_+_6543674 | 1.24 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr11_+_3959495 | 1.22 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr13_-_9061944 | 1.20 |
ENSDART00000164186
ENSDART00000102051 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr9_-_11587070 | 1.19 |
ENSDART00000030995
|
umps
|
uridine monophosphate synthetase |
| chr6_-_39080630 | 1.19 |
ENSDART00000021520
ENSDART00000128308 |
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr19_-_32804535 | 1.18 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr22_+_6293563 | 1.18 |
ENSDART00000063416
|
rnasel2
|
ribonuclease like 2 |
| chr6_-_54078623 | 1.17 |
ENSDART00000154076
|
hyal1
|
hyaluronoglucosaminidase 1 |
| chr7_+_17973269 | 1.16 |
ENSDART00000079969
|
kcnk7
|
potassium channel, subfamily K, member 7 |
| chr17_+_654759 | 1.16 |
ENSDART00000193703
|
CABZ01079748.1
|
|
| chr10_+_575929 | 1.16 |
ENSDART00000129856
|
smad4a
|
SMAD family member 4a |
| chr2_+_30547018 | 1.15 |
ENSDART00000193747
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr5_-_8765428 | 1.15 |
ENSDART00000167793
|
mybbp1a
|
MYB binding protein (P160) 1a |
| chr23_+_36122058 | 1.14 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr22_+_5118361 | 1.14 |
ENSDART00000168371
ENSDART00000170222 ENSDART00000158846 |
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr18_-_5850683 | 1.13 |
ENSDART00000082087
|
nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr22_-_12745589 | 1.13 |
ENSDART00000136841
|
plcd4a
|
phospholipase C, delta 4a |
| chr5_+_68807170 | 1.13 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr22_-_28777557 | 1.12 |
ENSDART00000135214
ENSDART00000131761 ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr20_+_25625872 | 1.12 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr21_+_17768174 | 1.11 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr16_-_8520474 | 1.11 |
ENSDART00000137365
|
grb10b
|
growth factor receptor-bound protein 10b |
| chr13_-_9295890 | 1.10 |
ENSDART00000110680
|
HTRA2 (1 of many)
|
si:dkey-33c12.12 |
| chr8_+_25295160 | 1.10 |
ENSDART00000049793
|
gstm.1
|
glutathione S-transferase mu, tandem duplicate 1 |
| chr4_+_2620751 | 1.08 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr8_+_15277874 | 1.08 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr17_-_19534474 | 1.07 |
ENSDART00000192469
|
cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr11_+_6010177 | 1.07 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr1_-_19648227 | 1.07 |
ENSDART00000054574
|
polr1e
|
polymerase (RNA) I polypeptide E |
| chr7_-_38340674 | 1.07 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr12_+_27117609 | 1.07 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr19_+_31061718 | 1.07 |
ENSDART00000145971
|
sostdc1b
|
sclerostin domain containing 1b |
| chr24_+_36317544 | 1.06 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr12_-_20350629 | 1.05 |
ENSDART00000066384
|
hbbe2
|
hemoglobin beta embryonic-2 |
| chr5_-_54195084 | 1.05 |
ENSDART00000171811
|
grk1b
|
G protein-coupled receptor kinase 1 b |
| chr20_-_40725142 | 1.02 |
ENSDART00000181279
|
cx32.2
|
connexin 32.2 |
| chr21_-_11327830 | 1.02 |
ENSDART00000122331
|
rtkn2b
|
rhotekin 2b |
| chr21_-_22122312 | 1.02 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr9_+_12887491 | 1.01 |
ENSDART00000102386
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr2_-_48171112 | 1.01 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr13_+_48359573 | 0.98 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr17_+_36627099 | 0.97 |
ENSDART00000154104
|
impg1b
|
interphotoreceptor matrix proteoglycan 1b |
| chr3_+_28939759 | 0.96 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr16_+_41974438 | 0.96 |
ENSDART00000102771
|
si:ch211-225p5.8
|
si:ch211-225p5.8 |
| chr3_+_51660158 | 0.96 |
ENSDART00000155512
ENSDART00000171764 |
chmp6a
|
charged multivesicular body protein 6a |
| chr20_-_26531850 | 0.95 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr14_+_16151368 | 0.95 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr17_+_51682429 | 0.94 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr14_+_5861435 | 0.93 |
ENSDART00000041279
ENSDART00000147341 |
tubb4b
|
tubulin, beta 4B class IVb |
| chr25_-_14424406 | 0.93 |
ENSDART00000073609
|
prmt7
|
protein arginine methyltransferase 7 |
| chr3_+_58379450 | 0.92 |
ENSDART00000155759
|
sdr42e2
|
short chain dehydrogenase/reductase family 42E, member 2 |
| chr3_-_25377163 | 0.92 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr3_-_41791178 | 0.92 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr1_-_54972170 | 0.91 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr19_+_21820144 | 0.91 |
ENSDART00000181996
|
CU856623.1
|
|
| chr23_+_38159715 | 0.90 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr16_-_54455573 | 0.90 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr11_+_25485774 | 0.89 |
ENSDART00000026249
|
gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr25_+_3231812 | 0.88 |
ENSDART00000163647
|
metap2b
|
methionyl aminopeptidase 2b |
| chr21_-_17603182 | 0.88 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr10_+_29849977 | 0.87 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr21_-_30408775 | 0.85 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr4_-_38033800 | 0.84 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr6_+_54078703 | 0.84 |
ENSDART00000110845
|
fhit
|
fragile histidine triad gene |
| chr25_+_245438 | 0.82 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr10_+_35002786 | 0.82 |
ENSDART00000099552
|
exosc8
|
exosome component 8 |
| chr1_+_31674297 | 0.81 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr10_+_29850330 | 0.81 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr4_+_3455665 | 0.81 |
ENSDART00000058277
|
znf800b
|
zinc finger protein 800b |
| chr13_-_4223955 | 0.80 |
ENSDART00000113060
|
dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr5_-_33935396 | 0.79 |
ENSDART00000133578
|
si:dkeyp-72a4.1
|
si:dkeyp-72a4.1 |
| chr14_+_8638353 | 0.79 |
ENSDART00000163240
|
si:dkeyp-115e12.6
|
si:dkeyp-115e12.6 |
| chr3_+_23029934 | 0.78 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr7_-_12596727 | 0.78 |
ENSDART00000186413
|
adamtsl3
|
ADAMTS-like 3 |
| chr11_-_44194132 | 0.77 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr25_-_36248053 | 0.77 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T cells 3b |
| chr23_-_35547613 | 0.77 |
ENSDART00000122387
|
alkal2a
|
ALK and LTK ligand 2a |
| chr7_-_30779575 | 0.76 |
ENSDART00000004782
|
mphosph10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr10_+_428269 | 0.76 |
ENSDART00000140715
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr14_+_14836468 | 0.76 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr4_+_18824959 | 0.75 |
ENSDART00000146141
ENSDART00000040424 |
slc26a3.1
|
solute carrier family 26 (anion exchanger), member 3 |
| chr1_-_17570013 | 0.75 |
ENSDART00000146946
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr2_-_32386598 | 0.74 |
ENSDART00000145575
|
ubtfl
|
upstream binding transcription factor, like |
| chr8_-_13678415 | 0.74 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr5_-_16475374 | 0.74 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of myca+mych
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 1.9 | 5.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.7 | 6.6 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 1.1 | 4.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 1.1 | 3.3 | GO:0021985 | neurohypophysis development(GO:0021985) |
| 1.0 | 10.5 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.8 | 4.7 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.7 | 2.0 | GO:1902626 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.6 | 6.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.6 | 2.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.6 | 1.7 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.6 | 6.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.5 | 1.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.4 | 1.3 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.4 | 1.6 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 4.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.4 | 1.1 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.4 | 1.1 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.4 | 1.1 | GO:0042942 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.3 | 4.5 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.3 | 1.0 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 1.2 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.3 | 3.0 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.3 | 3.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.3 | 2.0 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.3 | 2.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 0.9 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.3 | 1.7 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.3 | 0.8 | GO:0071042 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.3 | 0.8 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.3 | 1.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 | 1.0 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.2 | 1.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 1.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.2 | 2.4 | GO:0072088 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.2 | 6.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 0.9 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.2 | 4.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 1.7 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.2 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 2.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 0.8 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 1.7 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 2.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.2 | 1.9 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.2 | 1.1 | GO:0055014 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 1.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 0.7 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.2 | 2.7 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.2 | 5.9 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 14.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.2 | 4.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.8 | GO:0015961 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 1.0 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 0.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.8 | GO:0070378 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 3.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 1.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.3 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.3 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.0 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.2 | GO:0009265 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 0.1 | 1.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 2.0 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 4.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 4.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.3 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 1.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 1.6 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.1 | 0.2 | GO:0071028 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.4 | GO:0061339 | establishment of apical/basal cell polarity(GO:0035089) establishment of epithelial cell apical/basal polarity(GO:0045198) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 5.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 3.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.7 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.1 | 0.7 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 1.1 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 1.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.8 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 1.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 1.1 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 1.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.1 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 1.0 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 1.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 1.1 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.6 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 1.2 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 2.1 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 1.7 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 1.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 2.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.9 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 11.1 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 2.8 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.3 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.2 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.6 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 3.9 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 1.5 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 1.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.5 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 2.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 3.3 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 1.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.4 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.9 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.4 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.0 | 10.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.8 | 5.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.6 | 3.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.5 | 4.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.4 | 6.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 6.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 1.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.3 | 4.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.3 | 3.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 1.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 0.8 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 0.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 0.9 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 0.6 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 1.0 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 2.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 11.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 1.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 1.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 4.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 3.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.4 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 5.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 2.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.5 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.3 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.7 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.4 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 11.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 1.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 1.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 4.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 16.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.3 | 9.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 1.2 | 6.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 1.1 | 4.5 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 1.0 | 10.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.9 | 6.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.8 | 3.3 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.5 | 4.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 1.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.4 | 1.6 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.4 | 1.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 2.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.3 | 2.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 2.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 1.0 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.3 | 2.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 0.9 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 0.9 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 0.8 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 0.7 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.2 | 1.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 1.0 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.2 | 1.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.2 | 4.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 1.5 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 5.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 6.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.5 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 0.8 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 12.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 2.0 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.4 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 4.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 2.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.8 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 15.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 3.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 1.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.7 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.2 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 3.6 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 2.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 3.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.1 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.8 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 4.5 | GO:0015405 | P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 6.5 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 4.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 1.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 7.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 4.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 2.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 6.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.8 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 16.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 6.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 6.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 3.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.3 | 4.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 1.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 1.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 18.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 3.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.9 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 2.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 5.0 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |