Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
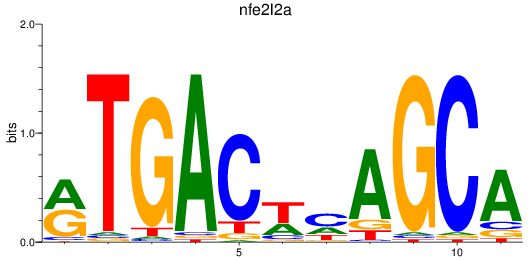
Results for nfe2l2a
Z-value: 0.87
Transcription factors associated with nfe2l2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfe2l2a
|
ENSDARG00000042824 | nuclear factor, erythroid 2-like 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfe2l2a | dr11_v1_chr9_+_1654284_1654284 | -0.11 | 3.0e-01 | Click! |
Activity profile of nfe2l2a motif
Sorted Z-values of nfe2l2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_7974155 | 14.21 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr23_-_15878879 | 12.57 |
ENSDART00000010119
|
eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr22_-_13857729 | 11.42 |
ENSDART00000177971
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr19_-_7450796 | 10.16 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr16_-_27640995 | 10.01 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr6_-_35446110 | 9.97 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr3_-_21280373 | 8.00 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr13_-_21688176 | 7.98 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr19_-_6385594 | 7.79 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr18_+_1703984 | 7.74 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr21_-_31143903 | 7.52 |
ENSDART00000111571
|
rap1gap2b
|
RAP1 GTPase activating protein 2b |
| chr8_+_24861264 | 7.50 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr17_+_27176243 | 7.46 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr24_-_24146875 | 7.24 |
ENSDART00000173052
|
map7d2b
|
MAP7 domain containing 2b |
| chr7_-_52842605 | 7.16 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr23_-_29376859 | 7.10 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr1_-_40227166 | 7.08 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr20_+_38724575 | 6.74 |
ENSDART00000015095
ENSDART00000152972 |
uts1
|
urotensin 1 |
| chr17_-_17948587 | 6.41 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr8_-_34052019 | 6.37 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr15_-_16070731 | 6.29 |
ENSDART00000122099
|
dynll2a
|
dynein, light chain, LC8-type 2a |
| chr5_-_31928913 | 6.10 |
ENSDART00000142919
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr23_+_19558574 | 6.04 |
ENSDART00000137811
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr24_+_41940299 | 5.80 |
ENSDART00000022349
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr1_-_53750522 | 5.75 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr5_-_71460556 | 5.65 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr6_+_21395051 | 5.30 |
ENSDART00000017774
|
cacng5a
|
calcium channel, voltage-dependent, gamma subunit 5a |
| chr11_+_45300669 | 5.17 |
ENSDART00000172238
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr20_-_39271844 | 5.13 |
ENSDART00000192708
|
clu
|
clusterin |
| chr16_-_17162485 | 5.12 |
ENSDART00000123011
|
iffo1b
|
intermediate filament family orphan 1b |
| chr23_-_18286822 | 4.90 |
ENSDART00000136672
|
fam19a1a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1a |
| chr20_-_39103119 | 4.70 |
ENSDART00000143379
|
rcan2
|
regulator of calcineurin 2 |
| chr24_+_24923166 | 4.64 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr2_-_9544161 | 4.53 |
ENSDART00000124425
|
slc25a24l
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24, like |
| chr24_+_26039464 | 4.34 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr11_-_3334248 | 4.31 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr19_-_17658160 | 4.30 |
ENSDART00000151766
ENSDART00000170790 ENSDART00000186678 ENSDART00000188045 ENSDART00000176980 ENSDART00000166313 ENSDART00000188589 |
thrb
|
thyroid hormone receptor beta |
| chr1_+_51312752 | 4.26 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr8_-_34051548 | 4.12 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr4_+_13696537 | 4.10 |
ENSDART00000109195
ENSDART00000122041 ENSDART00000192554 |
nrcama
|
neuronal cell adhesion molecule a |
| chr14_-_30490763 | 3.95 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr20_+_18661624 | 3.88 |
ENSDART00000152136
ENSDART00000126959 |
tnfaip2a
|
tumor necrosis factor, alpha-induced protein 2a |
| chr15_-_16679517 | 3.85 |
ENSDART00000177384
|
caln1
|
calneuron 1 |
| chr14_-_17588345 | 3.77 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr13_+_35746440 | 3.71 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr2_-_3038904 | 3.61 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr7_-_32833153 | 3.44 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr18_-_46763170 | 3.42 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr13_-_22907260 | 3.41 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr3_+_41731527 | 3.32 |
ENSDART00000049007
ENSDART00000187866 |
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr9_+_11202552 | 3.32 |
ENSDART00000151931
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr2_+_31437547 | 3.25 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr6_+_20651398 | 3.22 |
ENSDART00000183663
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr18_-_5692292 | 3.22 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr1_+_52137528 | 3.19 |
ENSDART00000007079
ENSDART00000074265 |
asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr10_+_38775408 | 3.12 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr5_-_50640171 | 3.11 |
ENSDART00000183353
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr14_+_34068075 | 3.09 |
ENSDART00000135556
|
lonrf1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr22_-_26323893 | 3.03 |
ENSDART00000105099
|
capn1b
|
calpain 1, (mu/I) large subunit b |
| chr21_+_9576176 | 3.03 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr18_-_41219790 | 2.98 |
ENSDART00000162387
ENSDART00000193753 |
zbtb38
|
zinc finger and BTB domain containing 38 |
| chr9_+_28688574 | 2.95 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr18_+_34478959 | 2.94 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr10_+_26747755 | 2.93 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr22_-_11829436 | 2.85 |
ENSDART00000126784
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr17_+_28533102 | 2.85 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr2_+_19236677 | 2.81 |
ENSDART00000166292
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr12_+_5209822 | 2.81 |
ENSDART00000152610
|
SLC35G1
|
si:ch211-197g18.2 |
| chr25_-_11378623 | 2.74 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr14_-_30490465 | 2.69 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr14_-_30960470 | 2.66 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr10_+_17235370 | 2.64 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr18_+_24922125 | 2.63 |
ENSDART00000180385
|
rgma
|
repulsive guidance molecule family member a |
| chr6_+_43903209 | 2.55 |
ENSDART00000006435
|
gpr27
|
G protein-coupled receptor 27 |
| chr10_+_29963518 | 2.45 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr16_-_30880236 | 2.44 |
ENSDART00000035583
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr2_-_26642831 | 2.41 |
ENSDART00000132854
ENSDART00000087714 ENSDART00000132651 |
u2surp
|
U2 snRNP-associated SURP domain containing |
| chr11_-_9948487 | 2.39 |
ENSDART00000189677
ENSDART00000113171 |
nlgn1
|
neuroligin 1 |
| chr7_+_63325819 | 2.33 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr20_+_26943072 | 2.32 |
ENSDART00000153215
|
cdca4
|
cell division cycle associated 4 |
| chr16_+_13855039 | 2.29 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr7_+_13582256 | 2.28 |
ENSDART00000158477
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr13_+_2357637 | 2.26 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr21_+_34686764 | 2.26 |
ENSDART00000005479
|
chm
|
choroideremia (Rab escort protein 1) |
| chr24_+_35911020 | 2.25 |
ENSDART00000088480
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr2_-_6115688 | 2.23 |
ENSDART00000081663
|
prdx1
|
peroxiredoxin 1 |
| chr21_-_40782393 | 2.22 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr13_+_1430094 | 2.22 |
ENSDART00000169888
|
si:ch211-165e15.1
|
si:ch211-165e15.1 |
| chr21_-_929293 | 2.22 |
ENSDART00000006419
|
txnl1
|
thioredoxin-like 1 |
| chr1_+_54737353 | 2.21 |
ENSDART00000130675
ENSDART00000162075 |
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr2_+_19236969 | 2.20 |
ENSDART00000163875
ENSDART00000168644 |
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr14_-_25985698 | 2.15 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr3_+_36160979 | 2.15 |
ENSDART00000038525
|
si:ch211-234h8.7
|
si:ch211-234h8.7 |
| chr19_+_31404686 | 2.15 |
ENSDART00000078459
|
pip4p2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr3_+_50201240 | 2.12 |
ENSDART00000156347
|
epn3a
|
epsin 3a |
| chr1_+_41690402 | 2.12 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr7_-_40630698 | 2.08 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr1_+_41609676 | 2.07 |
ENSDART00000183675
|
mogs
|
mannosyl-oligosaccharide glucosidase |
| chr21_-_929448 | 2.04 |
ENSDART00000133976
|
txnl1
|
thioredoxin-like 1 |
| chr2_+_37134281 | 2.02 |
ENSDART00000020135
|
pex19
|
peroxisomal biogenesis factor 19 |
| chr17_-_25737452 | 1.99 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr22_-_16416882 | 1.98 |
ENSDART00000062749
|
cts12
|
cathepsin 12 |
| chr13_+_24519175 | 1.95 |
ENSDART00000190790
|
sertad2b
|
SERTA domain containing 2b |
| chr7_-_23971497 | 1.93 |
ENSDART00000173603
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr10_-_4375190 | 1.92 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr23_-_12906228 | 1.89 |
ENSDART00000138807
|
ndnl2
|
necdin-like 2 |
| chr23_+_9508538 | 1.88 |
ENSDART00000010697
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr2_-_37134169 | 1.88 |
ENSDART00000146123
ENSDART00000146533 ENSDART00000040427 |
elavl1a
|
ELAV like RNA binding protein 1a |
| chr4_-_2052687 | 1.86 |
ENSDART00000138291
ENSDART00000150844 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr9_-_23253870 | 1.83 |
ENSDART00000143657
ENSDART00000169911 |
acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr2_+_9821757 | 1.81 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr2_-_6115389 | 1.81 |
ENSDART00000134921
|
prdx1
|
peroxiredoxin 1 |
| chr10_+_29265463 | 1.79 |
ENSDART00000155390
|
crebzf
|
CREB/ATF bZIP transcription factor |
| chr3_+_17846890 | 1.77 |
ENSDART00000193384
|
znf385c
|
zinc finger protein 385C |
| chr14_-_2004291 | 1.76 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr4_+_40218600 | 1.75 |
ENSDART00000152091
|
znf995
|
zinc finger protein 995 |
| chr11_-_23501467 | 1.75 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr3_+_32832042 | 1.74 |
ENSDART00000132679
ENSDART00000035759 |
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr11_+_24002503 | 1.73 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr18_-_3552414 | 1.70 |
ENSDART00000163762
ENSDART00000165434 ENSDART00000161197 ENSDART00000166841 ENSDART00000170260 |
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr8_-_18516385 | 1.69 |
ENSDART00000149446
|
si:dkey-30h22.11
|
si:dkey-30h22.11 |
| chr8_+_48942470 | 1.68 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr5_+_31959954 | 1.66 |
ENSDART00000142826
|
myo1hb
|
myosin IHb |
| chr23_+_32406009 | 1.64 |
ENSDART00000155793
|
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr23_-_15916316 | 1.64 |
ENSDART00000134096
ENSDART00000042469 ENSDART00000146605 |
mrgbp
|
MRG/MORF4L binding protein |
| chr20_+_48100261 | 1.63 |
ENSDART00000158604
|
xkr5a
|
XK related 5a |
| chr23_+_23183449 | 1.63 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr4_-_39111612 | 1.62 |
ENSDART00000150394
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr3_+_52684556 | 1.59 |
ENSDART00000139037
|
pgls
|
6-phosphogluconolactonase |
| chr18_-_19405616 | 1.57 |
ENSDART00000191290
ENSDART00000090855 |
megf11
|
multiple EGF-like-domains 11 |
| chr15_+_45591669 | 1.52 |
ENSDART00000157459
|
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr3_+_43374571 | 1.48 |
ENSDART00000182497
|
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr2_-_2813259 | 1.46 |
ENSDART00000032540
|
usp14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr24_-_9689915 | 1.46 |
ENSDART00000185972
ENSDART00000093046 |
uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr2_-_31800521 | 1.44 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr1_+_17376922 | 1.43 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr7_+_13491452 | 1.43 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr12_+_1455147 | 1.38 |
ENSDART00000018752
|
cops3
|
COP9 signalosome subunit 3 |
| chr7_+_62004048 | 1.35 |
ENSDART00000181818
|
smim20
|
small integral membrane protein 20 |
| chr5_-_5669879 | 1.34 |
ENSDART00000191963
|
CABZ01075628.1
|
|
| chr9_+_41103127 | 1.34 |
ENSDART00000000250
|
slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr21_+_25092300 | 1.32 |
ENSDART00000168505
|
dixdc1b
|
DIX domain containing 1b |
| chr16_+_24721914 | 1.31 |
ENSDART00000109459
|
smg9
|
smg9 nonsense mediated mRNA decay factor |
| chr16_+_24721556 | 1.30 |
ENSDART00000155754
|
smg9
|
smg9 nonsense mediated mRNA decay factor |
| chr1_+_6640437 | 1.28 |
ENSDART00000147638
|
si:ch211-93g23.2
|
si:ch211-93g23.2 |
| chr17_+_15674052 | 1.28 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr13_-_23007813 | 1.27 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr15_+_29393519 | 1.27 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr13_+_18311410 | 1.27 |
ENSDART00000036718
ENSDART00000132073 |
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr8_-_22660678 | 1.26 |
ENSDART00000181258
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr16_-_41804606 | 1.24 |
ENSDART00000134718
|
psmc4
|
proteasome 26S subunit, ATPase 4 |
| chr15_+_40792667 | 1.23 |
ENSDART00000193186
ENSDART00000184559 |
fat3a
|
FAT atypical cadherin 3a |
| chr2_+_9822319 | 1.21 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr8_+_10835456 | 1.21 |
ENSDART00000151388
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr6_+_33885828 | 1.19 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr4_+_54225677 | 1.17 |
ENSDART00000166637
|
znf995
|
zinc finger protein 995 |
| chr3_-_30153242 | 1.16 |
ENSDART00000077089
|
nucb1
|
nucleobindin 1 |
| chr14_+_24840669 | 1.16 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr20_-_8443425 | 1.10 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr18_-_42333428 | 1.08 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr16_-_13613475 | 1.06 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr4_+_20085114 | 1.06 |
ENSDART00000186698
ENSDART00000188635 |
ppp6r2a
|
protein phosphatase 6, regulatory subunit 2a |
| chr3_-_32831429 | 1.06 |
ENSDART00000184932
|
zgc:153733
|
zgc:153733 |
| chr21_-_41369370 | 1.05 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr8_-_39884359 | 1.02 |
ENSDART00000131372
|
mlec
|
malectin |
| chr6_-_12135741 | 1.02 |
ENSDART00000155090
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr6_-_21534301 | 0.99 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr9_+_23224761 | 0.99 |
ENSDART00000142008
|
map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr8_+_48943009 | 0.98 |
ENSDART00000180763
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr3_-_30152836 | 0.94 |
ENSDART00000165920
|
nucb1
|
nucleobindin 1 |
| chr16_+_10557504 | 0.93 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr3_+_26064091 | 0.93 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr25_+_19710619 | 0.92 |
ENSDART00000156435
|
mxg
|
myxovirus (influenza virus) resistance G |
| chr1_+_35473219 | 0.90 |
ENSDART00000109678
ENSDART00000181635 |
usp38
|
ubiquitin specific peptidase 38 |
| chr25_-_16818978 | 0.90 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr3_+_62140077 | 0.87 |
ENSDART00000108945
|
GID4
|
GID complex subunit 4 homolog |
| chr9_-_12885201 | 0.82 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr22_-_18116635 | 0.78 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr3_+_19687217 | 0.77 |
ENSDART00000141937
|
tlk2
|
tousled-like kinase 2 |
| chr15_-_39820491 | 0.75 |
ENSDART00000097134
|
robo1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr12_-_13729263 | 0.71 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr16_+_11029762 | 0.69 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr12_-_1034383 | 0.69 |
ENSDART00000152455
ENSDART00000152346 |
polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr8_+_42629748 | 0.68 |
ENSDART00000075553
ENSDART00000135525 |
crybb2
|
crystallin, beta B2 |
| chr8_+_20438884 | 0.60 |
ENSDART00000016422
ENSDART00000133794 |
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr4_+_33012407 | 0.59 |
ENSDART00000151873
|
si:dkey-26h11.2
|
si:dkey-26h11.2 |
| chr12_-_19151708 | 0.58 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr21_-_39327223 | 0.57 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr23_+_21278948 | 0.56 |
ENSDART00000156701
ENSDART00000033970 |
ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr17_-_22001303 | 0.55 |
ENSDART00000122190
|
slc22a7b.2
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 2 |
| chr5_-_54424019 | 0.52 |
ENSDART00000169270
ENSDART00000164067 |
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr7_-_18712482 | 0.51 |
ENSDART00000173517
ENSDART00000122564 ENSDART00000173905 |
trmt10a
|
tRNA methyltransferase 10A |
| chr21_+_37436907 | 0.49 |
ENSDART00000182611
ENSDART00000076328 |
pgrmc1
|
progesterone receptor membrane component 1 |
| chr14_+_21755469 | 0.48 |
ENSDART00000186326
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr12_+_35587971 | 0.48 |
ENSDART00000166072
ENSDART00000181861 ENSDART00000158043 |
chmp6b
|
charged multivesicular body protein 6b |
| chr9_-_33749556 | 0.48 |
ENSDART00000149383
|
ndp
|
Norrie disease (pseudoglioma) |
| chr14_-_17072736 | 0.48 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr2_-_24962002 | 0.44 |
ENSDART00000132050
|
hltf
|
helicase-like transcription factor |
| chr4_+_11690923 | 0.41 |
ENSDART00000150624
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr14_+_31496543 | 0.40 |
ENSDART00000170683
|
phf6
|
PHD finger protein 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfe2l2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0015824 | proline transport(GO:0015824) |
| 2.0 | 10.2 | GO:0090199 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 1.1 | 3.4 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 1.1 | 3.2 | GO:0015884 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 1.1 | 4.3 | GO:0042706 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.9 | 2.8 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.7 | 5.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.7 | 2.9 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.6 | 2.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.6 | 1.9 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.6 | 5.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.5 | 2.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.5 | 4.6 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.5 | 2.0 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.4 | 3.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.4 | 7.8 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.4 | 4.0 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.4 | 7.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.4 | 3.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 2.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 2.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.4 | 3.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 1.3 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.3 | 6.6 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.3 | 5.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.3 | 2.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 1.8 | GO:0046874 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.3 | 1.5 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 2.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 1.5 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.3 | 4.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.3 | 7.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.3 | 2.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 0.7 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.2 | 2.4 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 1.2 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 11.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.2 | 0.5 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.2 | 2.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 0.5 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 1.4 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 1.7 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 1.6 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 2.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 1.1 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 1.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 1.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 6.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.5 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 9.4 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.1 | 1.7 | GO:0045116 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 6.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 3.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 2.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 0.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 2.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.7 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 4.3 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 6.0 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 5.1 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.1 | 0.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.4 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 2.5 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.1 | 0.2 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 1.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.3 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 2.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 5.4 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 1.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 3.3 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.1 | 14.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 6.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 4.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 8.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 3.0 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 1.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 2.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 1.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.9 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.5 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 1.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 4.5 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 1.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 4.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 7.0 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 3.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 2.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 2.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 1.3 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 2.3 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 1.6 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 12.0 | GO:0007017 | microtubule-based process(GO:0007017) |
| 0.0 | 11.8 | GO:0048666 | neuron development(GO:0048666) |
| 0.0 | 4.6 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.5 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 1.3 | GO:0006413 | translational initiation(GO:0006413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 1.0 | 6.0 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.9 | 14.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.7 | 2.6 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.6 | 1.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.6 | 2.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.5 | 5.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.5 | 6.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 8.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 3.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 2.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 12.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 6.3 | GO:0043186 | P granule(GO:0043186) |
| 0.2 | 3.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 1.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 3.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 5.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 15.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 1.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 3.7 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.1 | 4.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.6 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 10.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 2.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 7.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.9 | GO:0098844 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 6.2 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 5.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 4.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.1 | 3.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 1.1 | 4.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.9 | 4.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.8 | 4.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.8 | 7.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.7 | 3.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.6 | 16.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.6 | 4.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.6 | 2.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.4 | 2.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.4 | 3.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.4 | 2.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.3 | 2.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 2.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 3.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 1.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 3.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 2.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 3.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 1.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 1.3 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 5.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 2.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 2.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 0.5 | GO:0030975 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.2 | 1.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 6.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 2.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 2.9 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.5 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 10.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 6.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 3.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 2.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 8.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 13.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.8 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.7 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 2.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 5.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.0 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 2.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 5.3 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 3.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 7.1 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 1.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 8.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 5.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.6 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 2.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 23.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 4.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 3.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 3.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 6.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.3 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 11.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.9 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 4.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 5.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.0 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 2.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 8.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 11.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.4 | 3.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 1.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 3.1 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.2 | 2.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 3.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 3.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 4.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 5.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 4.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |