Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
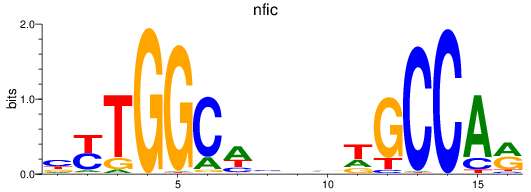
Results for nfic
Z-value: 0.53
Transcription factors associated with nfic
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfic
|
ENSDARG00000043210 | nuclear factor I/C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfic | dr11_v1_chr8_+_20679759_20679759 | 0.34 | 7.5e-04 | Click! |
Activity profile of nfic motif
Sorted Z-values of nfic motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_125091 | 11.48 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr1_+_7546259 | 11.37 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr3_+_31039923 | 6.52 |
ENSDART00000147706
|
cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr2_+_55982940 | 5.30 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr12_+_30653047 | 4.92 |
ENSDART00000148562
|
thbs2b
|
thrombospondin 2b |
| chr22_-_10487490 | 4.72 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr2_+_31665836 | 4.28 |
ENSDART00000135411
ENSDART00000143914 |
si:ch211-106h4.12
|
si:ch211-106h4.12 |
| chr14_+_15543331 | 3.89 |
ENSDART00000167025
|
si:dkey-203a12.7
|
si:dkey-203a12.7 |
| chr11_-_270210 | 3.87 |
ENSDART00000005217
ENSDART00000172779 |
alas1
|
aminolevulinate, delta-, synthase 1 |
| chr5_+_37517800 | 3.44 |
ENSDART00000048107
|
FP102018.1
|
Danio rerio latent transforming growth factor beta binding protein 3 (ltbp3), mRNA. |
| chr21_+_10866421 | 2.92 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr19_+_43780970 | 2.90 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr6_-_35401282 | 2.85 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr20_-_33976305 | 2.76 |
ENSDART00000111022
|
sele
|
selectin E |
| chr19_+_9786722 | 2.35 |
ENSDART00000138310
|
cacng6a
|
calcium channel, voltage-dependent, gamma subunit 6a |
| chr19_-_5332784 | 2.14 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr20_+_51464670 | 2.13 |
ENSDART00000150110
|
thbd
|
thrombomodulin |
| chr8_-_36399884 | 1.99 |
ENSDART00000108538
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr16_+_23397785 | 1.76 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr2_+_29257942 | 1.72 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr20_-_20410029 | 1.70 |
ENSDART00000192177
ENSDART00000063483 |
prkchb
|
protein kinase C, eta, b |
| chr9_+_37152564 | 1.54 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr14_+_30775066 | 1.52 |
ENSDART00000139975
ENSDART00000144921 |
atl3
|
atlastin 3 |
| chr25_-_36248053 | 1.50 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T cells 3b |
| chr15_-_18091157 | 1.48 |
ENSDART00000153848
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr1_+_33328857 | 1.42 |
ENSDART00000137151
|
mxra5a
|
matrix-remodelling associated 5a |
| chr3_-_40528333 | 1.35 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr1_+_49568335 | 1.33 |
ENSDART00000142957
|
col17a1a
|
collagen, type XVII, alpha 1a |
| chr10_-_8358396 | 1.33 |
ENSDART00000059322
|
csgalnact1a
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1a |
| chr21_+_38089036 | 1.31 |
ENSDART00000147219
|
klf8
|
Kruppel-like factor 8 |
| chr5_-_67629263 | 1.30 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr17_+_30546579 | 1.30 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr25_+_35058088 | 1.26 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr7_-_29341233 | 1.26 |
ENSDART00000140938
ENSDART00000147251 |
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr4_-_72179814 | 1.23 |
ENSDART00000172239
ENSDART00000189634 ENSDART00000186317 |
slco1f4
|
solute carrier organic anion transporter family, member 1F4 |
| chr6_-_13114406 | 1.20 |
ENSDART00000188015
|
zgc:194469
|
zgc:194469 |
| chr2_+_7818368 | 1.19 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr10_+_44042033 | 1.18 |
ENSDART00000190006
ENSDART00000046172 |
cryba4
|
crystallin, beta A4 |
| chr11_+_13630107 | 1.16 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr10_+_7636811 | 1.15 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr6_-_35032792 | 1.13 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr22_-_15693085 | 1.13 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr9_-_23884937 | 1.11 |
ENSDART00000141377
|
FAM124A
|
si:dkeyp-74a11.1 |
| chr4_-_6416155 | 1.10 |
ENSDART00000110535
|
MDFIC
|
si:ch73-156e19.1 |
| chr16_+_33143503 | 1.09 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr16_+_30604387 | 1.09 |
ENSDART00000058785
|
fam210ab
|
family with sequence similarity 210, member Ab |
| chr6_-_39199070 | 1.08 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr23_-_20363261 | 1.07 |
ENSDART00000054651
|
si:rp71-17i16.5
|
si:rp71-17i16.5 |
| chr19_-_9786914 | 1.05 |
ENSDART00000181669
|
si:dkey-14o18.2
|
si:dkey-14o18.2 |
| chr25_-_27564590 | 1.00 |
ENSDART00000073508
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr4_+_12292274 | 0.99 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr15_+_31806303 | 0.98 |
ENSDART00000155902
|
frya
|
furry homolog a (Drosophila) |
| chr20_+_23960525 | 0.96 |
ENSDART00000042123
|
cx52.6
|
connexin 52.6 |
| chr8_+_25616946 | 0.95 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr10_-_9115383 | 0.94 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr20_+_48739154 | 0.94 |
ENSDART00000100262
|
zgc:110348
|
zgc:110348 |
| chr6_-_6254432 | 0.94 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr13_+_18371208 | 0.93 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr12_+_4160804 | 0.92 |
ENSDART00000152515
|
itgam
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr16_+_21426524 | 0.91 |
ENSDART00000182869
|
gsdmeb
|
gasdermin Eb |
| chr15_-_14469704 | 0.90 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr4_-_6416414 | 0.89 |
ENSDART00000191136
|
MDFIC
|
si:ch73-156e19.1 |
| chr2_-_25159309 | 0.86 |
ENSDART00000137290
|
si:dkey-223d7.6
|
si:dkey-223d7.6 |
| chr25_-_29074064 | 0.85 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr7_-_26306546 | 0.82 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr12_+_30168342 | 0.81 |
ENSDART00000142756
|
ablim1b
|
actin binding LIM protein 1b |
| chr15_+_21672281 | 0.81 |
ENSDART00000153923
|
si:dkey-40g16.5
|
si:dkey-40g16.5 |
| chr20_-_8110672 | 0.81 |
ENSDART00000113993
|
si:ch211-232i5.1
|
si:ch211-232i5.1 |
| chr21_+_5257018 | 0.81 |
ENSDART00000183100
ENSDART00000191525 |
loxhd1a
|
lipoxygenase homology domains 1a |
| chr2_-_51303937 | 0.79 |
ENSDART00000164111
|
si:ch211-215e19.8
|
si:ch211-215e19.8 |
| chr2_+_6303639 | 0.78 |
ENSDART00000132859
|
otol1a
|
otolin 1a |
| chr16_-_43041324 | 0.78 |
ENSDART00000155445
ENSDART00000156836 ENSDART00000154945 |
si:dkey-7j14.6
|
si:dkey-7j14.6 |
| chr14_+_15484544 | 0.78 |
ENSDART00000188649
|
CR382326.1
|
|
| chr9_-_23885220 | 0.78 |
ENSDART00000112058
|
FAM124A
|
si:dkeyp-74a11.1 |
| chr23_-_26227805 | 0.75 |
ENSDART00000158082
|
BX927204.1
|
|
| chr8_+_43852743 | 0.74 |
ENSDART00000186485
|
AL808129.2
|
|
| chr3_-_55139127 | 0.73 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr9_-_39968820 | 0.70 |
ENSDART00000100311
|
IKZF2
|
si:zfos-1425h8.1 |
| chr10_+_36345176 | 0.68 |
ENSDART00000099397
|
or105-1
|
odorant receptor, family C, subfamily 105, member 1 |
| chr5_-_37935607 | 0.64 |
ENSDART00000141233
ENSDART00000084868 ENSDART00000125857 |
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr15_-_18200358 | 0.62 |
ENSDART00000158569
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr18_+_33818577 | 0.61 |
ENSDART00000131788
|
olfcq19
|
olfactory receptor C family, q19 |
| chr7_-_55675617 | 0.61 |
ENSDART00000021009
ENSDART00000188631 |
cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr23_+_4324625 | 0.61 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr5_+_55225497 | 0.60 |
ENSDART00000144087
|
tmc2a
|
transmembrane channel-like 2a |
| chr7_-_26306954 | 0.59 |
ENSDART00000057288
|
zgc:77439
|
zgc:77439 |
| chr23_+_25822742 | 0.58 |
ENSDART00000184436
|
r3hdml
|
R3H domain containing-like |
| chr13_+_40635844 | 0.56 |
ENSDART00000137310
|
hpse2
|
heparanase 2 |
| chr21_+_38634296 | 0.55 |
ENSDART00000114467
|
AL935199.1
|
|
| chr21_-_22681534 | 0.54 |
ENSDART00000159233
|
gig2f
|
grass carp reovirus (GCRV)-induced gene 2f |
| chr25_-_17367578 | 0.54 |
ENSDART00000064591
ENSDART00000110692 |
cyp2x6
|
cytochrome P450, family 2, subfamily X, polypeptide 6 |
| chr19_+_14109348 | 0.53 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr2_+_16798923 | 0.51 |
ENSDART00000181603
ENSDART00000179816 ENSDART00000087107 ENSDART00000187009 |
eif4g1a
|
eukaryotic translation initiation factor 4 gamma, 1a |
| chr5_-_37044262 | 0.49 |
ENSDART00000166670
|
si:dkeyp-110c7.8
|
si:dkeyp-110c7.8 |
| chr6_+_19933763 | 0.48 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr2_-_51259371 | 0.48 |
ENSDART00000158049
ENSDART00000164916 ENSDART00000157792 |
si:ch211-215e19.6
|
si:ch211-215e19.6 |
| chr6_-_34006917 | 0.47 |
ENSDART00000190702
ENSDART00000184003 |
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr4_+_59589201 | 0.47 |
ENSDART00000150576
|
si:dkey-4e4.1
|
si:dkey-4e4.1 |
| chr4_-_75899294 | 0.46 |
ENSDART00000157887
|
si:dkey-261j11.3
|
si:dkey-261j11.3 |
| chr8_-_25033681 | 0.46 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr4_-_38033800 | 0.46 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr16_+_35401543 | 0.43 |
ENSDART00000171608
|
rab42b
|
RAB42, member RAS oncogene family |
| chr17_-_50050453 | 0.43 |
ENSDART00000182057
|
zgc:100951
|
zgc:100951 |
| chr9_-_48937089 | 0.41 |
ENSDART00000193442
|
cers6
|
ceramide synthase 6 |
| chr7_+_9008372 | 0.41 |
ENSDART00000144897
|
AL935128.1
|
|
| chr5_-_57898008 | 0.40 |
ENSDART00000050945
|
layna
|
layilin a |
| chr9_-_46382637 | 0.40 |
ENSDART00000085738
|
lct
|
lactase |
| chr16_-_12488365 | 0.39 |
ENSDART00000172750
|
slc2a3b
|
solute carrier family 2 (facilitated glucose transporter), member 3b |
| chr3_+_57038033 | 0.36 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr4_+_61171310 | 0.36 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr25_+_4660639 | 0.35 |
ENSDART00000130299
|
deaf1
|
DEAF1 transcription factor |
| chr24_-_31942656 | 0.34 |
ENSDART00000180308
ENSDART00000016879 |
c1ql3a
|
complement component 1, q subcomponent-like 3a |
| chr5_-_32338866 | 0.33 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr13_+_33651416 | 0.32 |
ENSDART00000180221
|
BX005372.1
|
|
| chr11_+_14284866 | 0.29 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr23_-_45504991 | 0.29 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr11_-_43948885 | 0.29 |
ENSDART00000164522
|
CABZ01074203.1
|
|
| chr20_+_25568694 | 0.29 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr5_+_22791686 | 0.27 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr21_-_22678195 | 0.27 |
ENSDART00000171231
|
gig2g
|
grass carp reovirus (GCRV)-induced gene 2g |
| chr19_+_7636941 | 0.26 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr8_-_16515127 | 0.25 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr13_+_22731356 | 0.24 |
ENSDART00000133064
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr3_-_43646733 | 0.22 |
ENSDART00000180959
|
axin1
|
axin 1 |
| chr9_-_32142311 | 0.21 |
ENSDART00000142768
|
ankrd44
|
ankyrin repeat domain 44 |
| chr4_+_30718997 | 0.21 |
ENSDART00000190901
|
CU207245.1
|
|
| chr6_+_41186320 | 0.20 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr25_-_36261836 | 0.18 |
ENSDART00000179411
|
dus2
|
dihydrouridine synthase 2 |
| chr1_-_7659870 | 0.17 |
ENSDART00000085203
|
efnb2b
|
ephrin-B2b |
| chr7_+_73447724 | 0.17 |
ENSDART00000040786
|
cbln6
|
cerebellin 6 |
| chr3_+_22905341 | 0.17 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr14_+_8174828 | 0.15 |
ENSDART00000167228
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr10_-_43844537 | 0.15 |
ENSDART00000114208
|
tlr8b
|
toll-like receptor 8b |
| chr14_+_30774894 | 0.13 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr3_-_34279109 | 0.11 |
ENSDART00000183255
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr14_+_20893065 | 0.09 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr10_-_16065185 | 0.09 |
ENSDART00000187266
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr2_-_30912922 | 0.06 |
ENSDART00000141669
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr1_-_44434707 | 0.04 |
ENSDART00000110148
|
cryba1l2
|
crystallin, beta A1, like 2 |
| chr4_+_1283068 | 0.03 |
ENSDART00000167233
|
chrm2a
|
cholinergic receptor, muscarinic 2a |
| chr13_-_22774491 | 0.03 |
ENSDART00000189320
|
si:ch211-150i13.1
|
si:ch211-150i13.1 |
| chr7_+_36898622 | 0.02 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr15_+_24749985 | 0.01 |
ENSDART00000146107
|
ywhae1
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 1 |
| chr18_+_9493720 | 0.00 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfic
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.4 | 5.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 1.5 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.3 | 6.8 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 0.9 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.2 | 1.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.9 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.2 | 1.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 1.3 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 4.9 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 2.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.6 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 1.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.5 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 1.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.2 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 1.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 2.9 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 1.1 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.2 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 1.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.3 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 2.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 5.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 2.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 1.4 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.1 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.6 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 1.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 2.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.0 | GO:0030427 | site of polarized growth(GO:0030427) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.8 | 5.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.3 | 1.4 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 5.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 5.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 2.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.8 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.0 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.7 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 3.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 11.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 2.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 1.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 2.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |