Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
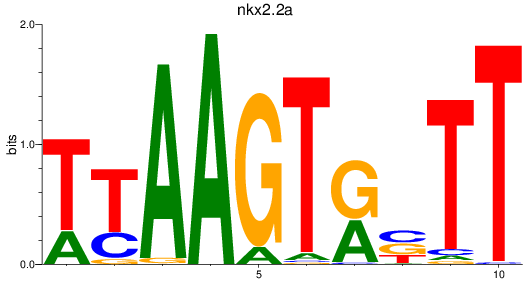
Results for nkx2.2a
Z-value: 0.64
Transcription factors associated with nkx2.2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2a
|
ENSDARG00000053298 | NK2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2a | dr11_v1_chr17_-_42213822_42213822 | 0.21 | 4.5e-02 | Click! |
Activity profile of nkx2.2a motif
Sorted Z-values of nkx2.2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_3629201 | 5.11 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr25_-_7974494 | 3.99 |
ENSDART00000171446
|
hal
|
histidine ammonia-lyase |
| chr19_+_9344171 | 3.67 |
ENSDART00000133447
ENSDART00000104622 |
si:ch211-288g17.4
|
si:ch211-288g17.4 |
| chr7_+_41295974 | 3.64 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr8_-_39739627 | 3.15 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr8_-_39739056 | 3.06 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr24_-_2829049 | 2.84 |
ENSDART00000164913
|
PPP1R3G
|
si:ch211-152c8.5 |
| chr2_+_1486822 | 2.81 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr2_+_1487118 | 2.74 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr20_+_53577502 | 2.70 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr17_-_14726824 | 2.70 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr16_-_25400257 | 2.67 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr13_-_18637244 | 2.58 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr22_-_24757785 | 2.52 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr2_-_20052561 | 2.36 |
ENSDART00000100133
|
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr7_-_69636502 | 2.22 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr13_+_47710434 | 2.12 |
ENSDART00000188724
|
TMEM87B
|
transmembrane protein 87B |
| chr13_-_40726865 | 2.06 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr20_-_6812688 | 2.03 |
ENSDART00000170934
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr21_+_9628854 | 1.98 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr5_+_31965845 | 1.98 |
ENSDART00000112968
|
myo1hb
|
myosin IHb |
| chr1_-_23294753 | 1.94 |
ENSDART00000013263
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr3_+_12829566 | 1.93 |
ENSDART00000157672
|
si:ch211-8c17.2
|
si:ch211-8c17.2 |
| chr14_-_42997145 | 1.92 |
ENSDART00000172801
|
pcdh10b
|
protocadherin 10b |
| chr1_+_45323142 | 1.89 |
ENSDART00000132210
|
emp1
|
epithelial membrane protein 1 |
| chr25_-_32311048 | 1.88 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr12_-_19865585 | 1.85 |
ENSDART00000066386
|
shisa9a
|
shisa family member 9a |
| chr8_+_31248917 | 1.81 |
ENSDART00000112170
|
unm_hu7912
|
un-named hu7912 |
| chr14_-_24110251 | 1.77 |
ENSDART00000079226
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr6_+_10338554 | 1.77 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr14_-_24110062 | 1.77 |
ENSDART00000177062
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr1_+_15137901 | 1.73 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr10_+_21807497 | 1.73 |
ENSDART00000164634
|
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr10_+_34394454 | 1.72 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr24_-_7699356 | 1.72 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr1_+_37196106 | 1.72 |
ENSDART00000008756
ENSDART00000157503 ENSDART00000162971 ENSDART00000191004 ENSDART00000078206 ENSDART00000045111 |
dclk2a
|
doublecortin-like kinase 2a |
| chr9_+_21722733 | 1.67 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr6_+_20650003 | 1.67 |
ENSDART00000049465
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr19_+_30867845 | 1.67 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr15_-_44601331 | 1.66 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr4_-_72609735 | 1.62 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr19_+_1184878 | 1.58 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr15_+_28106498 | 1.58 |
ENSDART00000041707
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr21_-_42055872 | 1.58 |
ENSDART00000144767
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr13_+_29352842 | 1.56 |
ENSDART00000113009
|
ogdhl
|
oxoglutarate dehydrogenase like |
| chr16_-_41465542 | 1.55 |
ENSDART00000169116
ENSDART00000187446 |
cpne4a
|
copine IVa |
| chr23_+_6795531 | 1.53 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr21_+_39948300 | 1.53 |
ENSDART00000137740
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr24_-_31332087 | 1.53 |
ENSDART00000161179
|
abcd3a
|
ATP-binding cassette, sub-family D (ALD), member 3a |
| chr23_-_27701361 | 1.51 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr25_+_35375848 | 1.51 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr24_-_33308045 | 1.50 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr5_+_72087619 | 1.47 |
ENSDART00000062885
|
oxt
|
oxytocin |
| chr7_+_73447724 | 1.47 |
ENSDART00000040786
|
cbln6
|
cerebellin 6 |
| chr9_-_9998087 | 1.45 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr19_+_37848830 | 1.44 |
ENSDART00000042276
ENSDART00000180872 |
nxph1
|
neurexophilin 1 |
| chr15_+_24691088 | 1.44 |
ENSDART00000110618
|
LRRC75A
|
si:dkey-151p21.7 |
| chr11_+_42422371 | 1.44 |
ENSDART00000163780
|
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr12_+_36413886 | 1.40 |
ENSDART00000126325
|
si:ch211-250n8.1
|
si:ch211-250n8.1 |
| chr1_-_45888608 | 1.39 |
ENSDART00000139219
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr4_+_37992753 | 1.38 |
ENSDART00000193890
|
CR759843.2
|
|
| chr6_+_10094061 | 1.38 |
ENSDART00000150998
ENSDART00000162236 |
atp7b
|
ATPase copper transporting beta |
| chr19_-_32940040 | 1.37 |
ENSDART00000179947
|
azin1b
|
antizyme inhibitor 1b |
| chr18_-_40481028 | 1.37 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr1_+_17695426 | 1.32 |
ENSDART00000103236
|
ankrd37
|
ankyrin repeat domain 37 |
| chr8_-_14484599 | 1.30 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr23_+_4362463 | 1.30 |
ENSDART00000039829
|
zgc:112175
|
zgc:112175 |
| chr24_+_22039964 | 1.29 |
ENSDART00000081220
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr16_-_31284922 | 1.29 |
ENSDART00000142638
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr24_+_12200260 | 1.27 |
ENSDART00000181391
|
CR855257.2
|
|
| chr14_-_24110707 | 1.27 |
ENSDART00000133522
ENSDART00000123152 |
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr2_-_24270062 | 1.24 |
ENSDART00000192445
|
myh7
|
myosin heavy chain 7 |
| chr16_-_12097394 | 1.24 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr8_+_28065803 | 1.23 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr13_-_29424454 | 1.23 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr15_-_20731297 | 1.23 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr13_-_24379271 | 1.23 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr14_-_25985698 | 1.23 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr2_-_37280028 | 1.23 |
ENSDART00000139459
|
nadkb
|
NAD kinase b |
| chr8_+_8845932 | 1.21 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr9_-_51323545 | 1.20 |
ENSDART00000139316
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr5_+_9408901 | 1.18 |
ENSDART00000193364
|
FP236810.1
|
|
| chr6_+_49255706 | 1.18 |
ENSDART00000156866
|
si:dkey-183k8.2
|
si:dkey-183k8.2 |
| chr20_+_32756763 | 1.17 |
ENSDART00000023006
|
fam84a
|
family with sequence similarity 84, member A |
| chr5_+_32009080 | 1.15 |
ENSDART00000186885
|
scai
|
suppressor of cancer cell invasion |
| chr11_-_29623380 | 1.15 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr20_-_28349144 | 1.14 |
ENSDART00000179690
ENSDART00000188059 |
ino80
|
INO80 complex subunit |
| chr19_+_24039830 | 1.14 |
ENSDART00000100422
|
rit1
|
Ras-like without CAAX 1 |
| chr5_-_60159116 | 1.14 |
ENSDART00000147675
|
si:dkey-280e8.1
|
si:dkey-280e8.1 |
| chr19_-_677713 | 1.13 |
ENSDART00000025146
|
slc6a19a.1
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 1 |
| chr6_+_9793791 | 1.13 |
ENSDART00000149896
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr21_-_25573064 | 1.12 |
ENSDART00000134310
|
CR388166.1
|
|
| chr2_+_38924975 | 1.12 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr5_+_54685175 | 1.11 |
ENSDART00000115016
|
pmchl
|
pro-melanin-concentrating hormone, like |
| chr7_-_26518086 | 1.11 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr4_+_19535946 | 1.11 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr23_+_6795709 | 1.11 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr4_-_18436899 | 1.10 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr16_+_17763848 | 1.10 |
ENSDART00000149408
ENSDART00000148878 |
them4
|
thioesterase superfamily member 4 |
| chr6_-_10168822 | 1.10 |
ENSDART00000151016
|
b3galt1a
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1a |
| chr21_+_11684830 | 1.09 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr17_+_30205258 | 1.08 |
ENSDART00000076596
ENSDART00000153795 |
spata17
|
spermatogenesis associated 17 |
| chr3_+_24134418 | 1.05 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr13_+_33368140 | 1.05 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr2_-_11662851 | 1.03 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr1_-_625875 | 1.03 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr2_+_33052169 | 1.03 |
ENSDART00000180008
|
rnf220a
|
ring finger protein 220a |
| chr3_+_49521106 | 1.02 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr20_+_27020201 | 1.02 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr24_+_5935377 | 1.01 |
ENSDART00000191989
ENSDART00000185932 ENSDART00000131768 |
abi1a
|
abl-interactor 1a |
| chr12_+_16281312 | 0.99 |
ENSDART00000152500
|
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr23_-_24856025 | 0.99 |
ENSDART00000142171
|
syt6a
|
synaptotagmin VIa |
| chr25_-_30047477 | 0.98 |
ENSDART00000164859
|
CR759810.1
|
|
| chr2_+_22531185 | 0.98 |
ENSDART00000171959
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr13_+_28675686 | 0.98 |
ENSDART00000027213
|
inaa
|
internexin neuronal intermediate filament protein, alpha a |
| chrM_+_12897 | 0.96 |
ENSDART00000093622
|
mt-nd5
|
NADH dehydrogenase 5, mitochondrial |
| chr15_+_19472129 | 0.95 |
ENSDART00000154779
|
zgc:77784
|
zgc:77784 |
| chr25_+_36152215 | 0.95 |
ENSDART00000036147
|
irx5b
|
iroquois homeobox 5b |
| chr2_+_7557912 | 0.94 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr16_-_22225295 | 0.94 |
ENSDART00000163519
|
LO017682.1
|
|
| chr21_+_11685009 | 0.93 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr9_-_23156908 | 0.92 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr13_-_23270576 | 0.92 |
ENSDART00000132828
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr18_-_14937211 | 0.92 |
ENSDART00000141893
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr11_-_3954691 | 0.92 |
ENSDART00000182041
|
pbrm1
|
polybromo 1 |
| chr8_+_14058646 | 0.90 |
ENSDART00000080852
|
ugt5e1
|
UDP glucuronosyltransferase 5 family, polypeptide E1 |
| chr6_+_4872883 | 0.90 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr17_+_10501647 | 0.90 |
ENSDART00000140391
|
tyro3
|
TYRO3 protein tyrosine kinase |
| chr23_-_39784368 | 0.89 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr19_-_22328154 | 0.89 |
ENSDART00000090464
|
si:ch73-196l6.5
|
si:ch73-196l6.5 |
| chr14_+_3449780 | 0.88 |
ENSDART00000163849
|
trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr3_+_40856095 | 0.88 |
ENSDART00000143207
|
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr23_+_36730713 | 0.88 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr15_-_13254480 | 0.87 |
ENSDART00000190499
|
zgc:172282
|
zgc:172282 |
| chr8_+_3431671 | 0.86 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr8_-_18262149 | 0.86 |
ENSDART00000143000
|
rnf220b
|
ring finger protein 220b |
| chr4_-_41269844 | 0.86 |
ENSDART00000186177
|
CR388165.2
|
|
| chr4_+_2230701 | 0.85 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr9_+_50600355 | 0.85 |
ENSDART00000187567
|
fign
|
fidgetin |
| chr13_-_32995324 | 0.85 |
ENSDART00000140542
ENSDART00000037740 |
kcnf1b
|
potassium voltage-gated channel, subfamily F, member 1b |
| chr23_+_40460333 | 0.85 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr8_-_1698155 | 0.84 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr4_+_13412030 | 0.83 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr2_+_31437547 | 0.82 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr5_+_20035284 | 0.81 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr4_+_18824959 | 0.81 |
ENSDART00000146141
ENSDART00000040424 |
slc26a3.1
|
solute carrier family 26 (anion exchanger), member 3 |
| chr14_-_49133426 | 0.81 |
ENSDART00000042421
|
ppp2ca
|
protein phosphatase 2, catalytic subunit, alpha isozyme |
| chr11_+_583142 | 0.80 |
ENSDART00000168157
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr16_-_11778933 | 0.80 |
ENSDART00000184916
|
pafah1b3
|
platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit |
| chr15_+_29292154 | 0.79 |
ENSDART00000137817
|
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr2_+_20472150 | 0.78 |
ENSDART00000168537
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr16_-_39267185 | 0.78 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr15_-_16070731 | 0.78 |
ENSDART00000122099
|
dynll2a
|
dynein, light chain, LC8-type 2a |
| chr11_+_18873113 | 0.76 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr9_-_22158784 | 0.76 |
ENSDART00000167850
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr5_+_32009542 | 0.76 |
ENSDART00000182025
ENSDART00000179879 |
scai
|
suppressor of cancer cell invasion |
| chr17_+_50074372 | 0.76 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr6_+_21395051 | 0.76 |
ENSDART00000017774
|
cacng5a
|
calcium channel, voltage-dependent, gamma subunit 5a |
| chr10_-_32851847 | 0.74 |
ENSDART00000134255
|
trim37
|
tripartite motif containing 37 |
| chr9_+_42095220 | 0.73 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr10_+_36441124 | 0.73 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr5_+_41143563 | 0.73 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr2_+_243778 | 0.73 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr2_+_2967255 | 0.72 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr17_-_30205302 | 0.72 |
ENSDART00000156827
|
si:dkey-27l15.1
|
si:dkey-27l15.1 |
| chr4_+_4267451 | 0.71 |
ENSDART00000192069
|
ano2
|
anoctamin 2 |
| chr10_+_5135842 | 0.71 |
ENSDART00000132627
ENSDART00000162434 |
zgc:113274
|
zgc:113274 |
| chr20_-_34750045 | 0.71 |
ENSDART00000186130
|
znf395b
|
zinc finger protein 395b |
| chr14_+_14043793 | 0.70 |
ENSDART00000164376
|
rraga
|
Ras-related GTP binding A |
| chr17_+_27723490 | 0.70 |
ENSDART00000123588
ENSDART00000170462 ENSDART00000169708 |
qkia
|
QKI, KH domain containing, RNA binding a |
| chr16_-_21181128 | 0.70 |
ENSDART00000133403
|
pde11al
|
phosphodiesterase 11a, like |
| chr8_-_13823091 | 0.70 |
ENSDART00000177174
ENSDART00000137021 |
cabp4
|
calcium binding protein 4 |
| chr7_-_35083585 | 0.69 |
ENSDART00000192732
|
agrp
|
agouti related neuropeptide |
| chr2_+_37424261 | 0.69 |
ENSDART00000132427
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr9_-_31596016 | 0.68 |
ENSDART00000142289
|
nalcn
|
sodium leak channel, non-selective |
| chr23_+_20513104 | 0.68 |
ENSDART00000079591
|
dpm1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr19_-_41213718 | 0.68 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr16_+_22654481 | 0.68 |
ENSDART00000179762
|
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr14_-_38889840 | 0.68 |
ENSDART00000035779
|
zgc:101583
|
zgc:101583 |
| chr6_-_39218609 | 0.67 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr1_-_51261420 | 0.67 |
ENSDART00000168685
|
kif16ba
|
kinesin family member 16Ba |
| chr18_+_32615075 | 0.67 |
ENSDART00000166937
|
CABZ01012885.1
|
|
| chr7_+_13527971 | 0.66 |
ENSDART00000020542
|
plekho2
|
pleckstrin homology domain containing, family O member 2 |
| chr16_+_813780 | 0.66 |
ENSDART00000162474
ENSDART00000161774 |
irx1a
|
iroquois homeobox 1a |
| chr4_-_27129697 | 0.65 |
ENSDART00000131240
|
zbed4
|
zinc finger, BED-type containing 4 |
| chr8_+_26292560 | 0.64 |
ENSDART00000053463
|
mgll
|
monoglyceride lipase |
| chr15_+_5339056 | 0.64 |
ENSDART00000174319
|
CABZ01018874.2
|
|
| chr11_+_18873619 | 0.64 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr14_+_46342882 | 0.63 |
ENSDART00000193707
ENSDART00000060577 |
tmem33
|
transmembrane protein 33 |
| chr24_-_21989406 | 0.63 |
ENSDART00000032963
|
apoob
|
apolipoprotein O, b |
| chr12_+_16440708 | 0.63 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr11_+_34522554 | 0.62 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr15_-_30714130 | 0.62 |
ENSDART00000156914
ENSDART00000154714 |
msi2b
|
musashi RNA-binding protein 2b |
| chr21_-_21790372 | 0.62 |
ENSDART00000151094
|
xrra1
|
X-ray radiation resistance associated 1 |
| chr7_-_40122139 | 0.61 |
ENSDART00000173982
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr6_+_36795225 | 0.61 |
ENSDART00000171504
|
si:ch73-29l19.1
|
si:ch73-29l19.1 |
| chr5_-_17601759 | 0.60 |
ENSDART00000138387
|
si:ch211-130h14.6
|
si:ch211-130h14.6 |
| chr10_-_5135788 | 0.60 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr20_+_33904258 | 0.60 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr12_-_35883814 | 0.59 |
ENSDART00000177986
ENSDART00000129888 |
cep131
|
centrosomal protein 131 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0019557 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.8 | 2.4 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.7 | 4.8 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.6 | 1.7 | GO:0051958 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.5 | 2.7 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.4 | 1.2 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.4 | 1.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.4 | 2.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.3 | 1.7 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.3 | 1.0 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.3 | 1.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.3 | 2.6 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.3 | 1.6 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.3 | 1.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.3 | 1.9 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.3 | 0.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 1.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 0.5 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.2 | 1.0 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.2 | 1.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.7 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.2 | 0.9 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.2 | 1.4 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 1.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.2 | 1.7 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 1.9 | GO:0021794 | thalamus development(GO:0021794) |
| 0.2 | 1.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.6 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 0.8 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.6 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 2.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 4.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 1.0 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 1.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 3.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 0.9 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.4 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) |
| 0.1 | 0.7 | GO:1990173 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 1.5 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.9 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.7 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 1.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 1.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.5 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 1.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.3 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.8 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.3 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) distal tubule morphogenesis(GO:0072156) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.1 | 1.4 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.9 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.8 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) negative regulation of intracellular protein transport(GO:0090317) |
| 0.1 | 0.3 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.3 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.6 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 1.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 | 0.5 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.7 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 2.5 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.5 | GO:0046070 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.4 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.9 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 0.2 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 0.8 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 2.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.9 | GO:0032885 | regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.0 | 1.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 1.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.8 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.6 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.4 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.5 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.6 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.4 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 2.0 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 2.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.3 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.4 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.6 | GO:0086010 | membrane depolarization(GO:0051899) membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.1 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 1.8 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.2 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.0 | 0.7 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 4.0 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0008584 | male gonad development(GO:0008584) |
| 0.0 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.6 | GO:0002068 | glandular epithelial cell development(GO:0002068) type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.7 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.3 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.4 | GO:0045841 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 2.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 2.2 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 2.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0021594 | rhombomere formation(GO:0021594) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 3.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 1.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 1.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 1.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.2 | 0.5 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.4 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.1 | 1.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.7 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.9 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 3.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 1.7 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.1 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.5 | GO:0001650 | fibrillar center(GO:0001650) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 0.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 6.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 3.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.9 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 4.8 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0036126 | sperm flagellum(GO:0036126) sperm part(GO:0097223) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 14.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.9 | 2.7 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.7 | 2.1 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.6 | 2.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.6 | 1.7 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.5 | 1.9 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.5 | 2.4 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.4 | 2.5 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.3 | 4.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 0.8 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 1.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.3 | 1.1 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.2 | 1.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 2.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 1.4 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 1.8 | GO:0001006 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.2 | 1.7 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 1.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 0.8 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.2 | 0.8 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 1.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 2.0 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.9 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.9 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 1.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.9 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 2.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 2.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.5 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.5 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 0.6 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.6 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.9 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.8 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.5 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.1 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 2.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 2.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 2.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.1 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.0 | 0.2 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.8 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 5.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 3.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 2.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.8 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 2.0 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.2 | 2.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 1.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 4.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 4.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 1.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.9 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 0.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |