Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
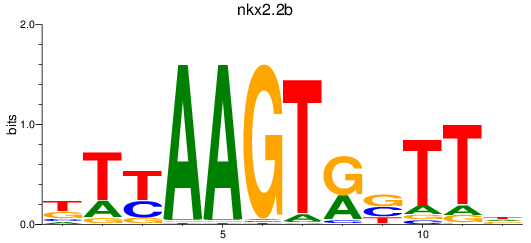
Results for nkx2.2b
Z-value: 0.61
Transcription factors associated with nkx2.2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2b
|
ENSDARG00000101549 | NK2 homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2b | dr11_v1_chr20_+_48782068_48782068 | -0.41 | 4.7e-05 | Click! |
Activity profile of nkx2.2b motif
Sorted Z-values of nkx2.2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_8840772 | 3.87 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr9_-_33063083 | 3.20 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr3_-_27647845 | 3.09 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr3_-_49110710 | 3.05 |
ENSDART00000160404
|
trim35-12
|
tripartite motif containing 35-12 |
| chr16_+_29586004 | 3.05 |
ENSDART00000149520
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr24_-_30263301 | 3.02 |
ENSDART00000162328
|
snx7
|
sorting nexin 7 |
| chr11_-_30636163 | 3.00 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr15_-_20468302 | 2.78 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr19_+_9344171 | 2.73 |
ENSDART00000133447
ENSDART00000104622 |
si:ch211-288g17.4
|
si:ch211-288g17.4 |
| chr5_-_38777852 | 2.73 |
ENSDART00000131603
|
si:dkey-58f10.4
|
si:dkey-58f10.4 |
| chr22_+_11775269 | 2.72 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr7_-_16562200 | 2.59 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr4_-_25215968 | 2.58 |
ENSDART00000066932
ENSDART00000066933 |
itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr15_+_13984879 | 2.56 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr2_-_28396993 | 2.55 |
ENSDART00000188170
|
CABZ01056052.1
|
|
| chr5_-_37117778 | 2.44 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr2_-_51772438 | 2.44 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr8_+_25302172 | 2.37 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr16_+_23913943 | 2.34 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr7_-_22790630 | 2.29 |
ENSDART00000173496
|
si:ch211-15b10.6
|
si:ch211-15b10.6 |
| chr5_+_1877464 | 2.28 |
ENSDART00000050658
|
zgc:101699
|
zgc:101699 |
| chr20_-_5267600 | 2.27 |
ENSDART00000099258
|
cyp46a1.4
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 4 |
| chr7_+_35268054 | 2.26 |
ENSDART00000113842
|
dpep2
|
dipeptidase 2 |
| chr12_-_48671612 | 2.22 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr9_-_9998087 | 2.21 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr8_-_38317914 | 2.20 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr8_-_50482781 | 2.15 |
ENSDART00000056361
|
ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr1_+_17900306 | 2.12 |
ENSDART00000089480
|
cyp4v8
|
cytochrome P450, family 4, subfamily V, polypeptide 8 |
| chr25_+_10416583 | 2.10 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr22_-_17631675 | 2.09 |
ENSDART00000132565
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr13_-_30996072 | 2.08 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr20_-_28349144 | 2.07 |
ENSDART00000179690
ENSDART00000188059 |
ino80
|
INO80 complex subunit |
| chr12_-_30558694 | 2.03 |
ENSDART00000153417
|
si:ch211-28p3.3
|
si:ch211-28p3.3 |
| chr4_+_2230701 | 2.03 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr12_-_22400999 | 2.02 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr2_+_48282590 | 2.01 |
ENSDART00000035338
|
lpar5a
|
lysophosphatidic acid receptor 5a |
| chr9_+_41156287 | 2.01 |
ENSDART00000189195
ENSDART00000186270 |
stat4
|
signal transducer and activator of transcription 4 |
| chr2_+_1487118 | 1.95 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr2_+_26498446 | 1.94 |
ENSDART00000078412
|
rps8a
|
ribosomal protein S8a |
| chr18_-_7631301 | 1.92 |
ENSDART00000062166
|
trim35-30
|
tripartite motif containing 35-30 |
| chr11_+_31285127 | 1.91 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr22_+_21317597 | 1.88 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr14_+_26759332 | 1.88 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr9_-_18911608 | 1.84 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr23_-_39784368 | 1.83 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr5_-_37900350 | 1.80 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr23_+_1078072 | 1.79 |
ENSDART00000159263
ENSDART00000053527 |
slc34a2b
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 2b |
| chr19_-_24555935 | 1.78 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr13_-_40401870 | 1.76 |
ENSDART00000128951
|
nkx3.3
|
NK3 homeobox 3 |
| chr2_-_37956768 | 1.76 |
ENSDART00000034595
|
cbln10
|
cerebellin 10 |
| chr23_+_38245610 | 1.74 |
ENSDART00000191386
|
znf217
|
zinc finger protein 217 |
| chr14_+_1170968 | 1.73 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr23_-_24394719 | 1.71 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr7_+_12950507 | 1.71 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr7_-_26518086 | 1.69 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr2_+_1486822 | 1.69 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr11_+_14286160 | 1.69 |
ENSDART00000166236
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr2_+_36114194 | 1.67 |
ENSDART00000113547
|
traj39
|
T-cell receptor alpha joining 39 |
| chr7_+_50849142 | 1.67 |
ENSDART00000073806
|
pcolceb
|
procollagen C-endopeptidase enhancer b |
| chr13_-_22699024 | 1.64 |
ENSDART00000016946
|
glud1a
|
glutamate dehydrogenase 1a |
| chr1_+_52560549 | 1.64 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr13_-_40237121 | 1.63 |
ENSDART00000145635
|
loxl4
|
lysyl oxidase-like 4 |
| chr1_-_47122058 | 1.63 |
ENSDART00000159925
ENSDART00000101143 ENSDART00000176803 |
mhc1zea
|
major histocompatibility complex class I ZEA |
| chr2_+_36121373 | 1.62 |
ENSDART00000187002
|
CT867973.2
|
|
| chr8_+_1170015 | 1.61 |
ENSDART00000081457
ENSDART00000164116 |
ccl27a
|
chemokine (C-C motif) ligand 27a |
| chr4_-_9891874 | 1.61 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr7_-_38087865 | 1.61 |
ENSDART00000052366
|
cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr1_-_43905252 | 1.58 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr23_+_36653376 | 1.58 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr19_-_24555623 | 1.58 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr22_+_883678 | 1.57 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr1_-_354115 | 1.57 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr23_+_20803270 | 1.56 |
ENSDART00000097381
|
zgc:154075
|
zgc:154075 |
| chr7_+_21275152 | 1.56 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr13_-_25819825 | 1.55 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr8_-_45760087 | 1.55 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr14_+_23717165 | 1.54 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr18_+_37272568 | 1.52 |
ENSDART00000132749
|
tmem123
|
transmembrane protein 123 |
| chr3_+_4403980 | 1.46 |
ENSDART00000156881
|
CR774195.1
|
|
| chr21_+_33454147 | 1.45 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr2_-_24269911 | 1.45 |
ENSDART00000099532
|
myh7
|
myosin heavy chain 7 |
| chr1_+_36651059 | 1.44 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr1_+_1915967 | 1.43 |
ENSDART00000131463
|
si:ch211-132g1.1
|
si:ch211-132g1.1 |
| chr12_-_28794957 | 1.42 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr19_-_48330287 | 1.42 |
ENSDART00000162244
|
si:ch73-359m17.7
|
si:ch73-359m17.7 |
| chr24_+_38205798 | 1.40 |
ENSDART00000141532
|
igl3v1
|
immunoglobulin light 3 variable 1 |
| chr15_-_1198886 | 1.40 |
ENSDART00000063285
|
lxn
|
latexin |
| chr17_+_6538733 | 1.40 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr12_-_30548244 | 1.40 |
ENSDART00000193616
|
zgc:158404
|
zgc:158404 |
| chr7_+_32901658 | 1.39 |
ENSDART00000115420
|
ano9b
|
anoctamin 9b |
| chr6_+_11438972 | 1.36 |
ENSDART00000029314
|
col5a2b
|
collagen, type V, alpha 2b |
| chr4_+_6032640 | 1.31 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr4_-_22311610 | 1.31 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr1_+_8534698 | 1.29 |
ENSDART00000021504
|
smcr8b
|
Smith-Magenis syndrome chromosome region, candidate 8b |
| chr22_-_7539414 | 1.29 |
ENSDART00000159083
|
BX511034.5
|
|
| chr20_+_33904258 | 1.28 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr3_+_19216567 | 1.27 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr13_+_44857087 | 1.24 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr9_+_18023288 | 1.24 |
ENSDART00000098355
|
tnfsf11
|
TNF superfamily member 11 |
| chr14_-_25956804 | 1.23 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr24_-_33308045 | 1.23 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr6_-_54107269 | 1.22 |
ENSDART00000190017
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr2_+_2967255 | 1.22 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr5_+_26795773 | 1.21 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr20_-_32045057 | 1.21 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr11_-_25257595 | 1.21 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr16_+_23087326 | 1.20 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr4_+_58576146 | 1.17 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr23_+_35847538 | 1.16 |
ENSDART00000143935
|
rarga
|
retinoic acid receptor gamma a |
| chr21_+_25793970 | 1.14 |
ENSDART00000101217
|
CLDN4 (1 of many)
|
zgc:136892 |
| chr6_-_41085443 | 1.13 |
ENSDART00000143944
ENSDART00000028217 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr18_-_50152689 | 1.13 |
ENSDART00000006078
|
loxl1
|
lysyl oxidase-like 1 |
| chr8_-_38105053 | 1.12 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr15_+_37589698 | 1.12 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr24_+_38197852 | 1.12 |
ENSDART00000142949
|
igl3v4
|
immunoglobulin light 3 variable 4 |
| chr8_-_43847138 | 1.12 |
ENSDART00000148358
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr22_-_6941098 | 1.11 |
ENSDART00000105864
|
zgc:171500
|
zgc:171500 |
| chr2_-_58183499 | 1.10 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr7_+_57108823 | 1.10 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr16_-_51288178 | 1.09 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr21_-_36619599 | 1.09 |
ENSDART00000065208
|
nop16
|
NOP16 nucleolar protein homolog (yeast) |
| chr11_+_3308656 | 1.09 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr21_+_19547806 | 1.07 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr17_+_20616196 | 1.07 |
ENSDART00000153515
|
si:ch73-306e8.2
|
si:ch73-306e8.2 |
| chr16_-_33105677 | 1.07 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr7_-_8309505 | 1.06 |
ENSDART00000182530
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr4_-_16644708 | 1.06 |
ENSDART00000042307
|
sinhcaf
|
SIN3-HDAC complex associated factor |
| chr6_-_41079209 | 1.06 |
ENSDART00000151592
|
rab44
|
RAB44, member RAS oncogene family |
| chr14_-_38873095 | 1.05 |
ENSDART00000047050
ENSDART00000173285 ENSDART00000147521 |
gsr
|
glutathione reductase |
| chr2_-_36494308 | 1.04 |
ENSDART00000110378
|
BX901889.2
|
|
| chr5_+_43782267 | 1.04 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr3_-_11008532 | 1.03 |
ENSDART00000165086
|
CR382337.3
|
|
| chr25_+_4635355 | 1.03 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr4_-_68901741 | 1.02 |
ENSDART00000163087
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr3_+_60761811 | 1.01 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr20_-_39333657 | 1.00 |
ENSDART00000153720
ENSDART00000142164 |
ccl38.1
|
chemokine (C-C motif) ligand 38, duplicate 1 |
| chr4_+_62184754 | 1.00 |
ENSDART00000168844
|
si:dkeyp-35e5.9
|
si:dkeyp-35e5.9 |
| chr23_-_27701361 | 0.99 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr23_+_19813677 | 0.99 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr19_+_4066449 | 0.99 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr1_+_55703120 | 0.97 |
ENSDART00000141089
|
adgre6
|
adhesion G protein-coupled receptor E6 |
| chr5_+_3927989 | 0.95 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr23_-_40295610 | 0.95 |
ENSDART00000186719
|
CABZ01067002.1
|
|
| chr9_+_1313418 | 0.95 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr19_-_808265 | 0.93 |
ENSDART00000082454
|
glmp
|
glycosylated lysosomal membrane protein |
| chr2_-_51757328 | 0.93 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr4_-_7673165 | 0.93 |
ENSDART00000028171
|
lta4h
|
leukotriene A4 hydrolase |
| chr3_-_16110100 | 0.92 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr12_+_33320884 | 0.92 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr24_-_26484298 | 0.92 |
ENSDART00000140647
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr9_+_40546677 | 0.92 |
ENSDART00000132911
|
vwc2l
|
von Willebrand factor C domain containing protein 2-like |
| chr3_+_16841942 | 0.91 |
ENSDART00000023985
ENSDART00000145317 |
stk17al
|
serine/threonine kinase 17a like |
| chr1_+_55723870 | 0.90 |
ENSDART00000146614
|
adgre17
|
adhesion G protein-coupled receptor E17 |
| chr2_-_7696287 | 0.90 |
ENSDART00000190769
|
CABZ01055306.1
|
|
| chr6_-_26895314 | 0.90 |
ENSDART00000134259
|
hdlbpa
|
high density lipoprotein binding protein a |
| chr2_-_1514001 | 0.90 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr16_+_35887868 | 0.90 |
ENSDART00000169677
ENSDART00000170772 |
thrap3a
|
thyroid hormone receptor associated protein 3a |
| chr18_+_40471826 | 0.89 |
ENSDART00000098806
|
ugt5c3
|
UDP glucuronosyltransferase 5 family, polypeptide C3 |
| chr11_+_37638873 | 0.89 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr25_+_15994100 | 0.88 |
ENSDART00000144723
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr4_-_75552652 | 0.88 |
ENSDART00000169253
ENSDART00000182360 |
si:dkey-71l4.4
si:dkey-71l4.5
|
si:dkey-71l4.4 si:dkey-71l4.5 |
| chr20_-_34750045 | 0.87 |
ENSDART00000186130
|
znf395b
|
zinc finger protein 395b |
| chr6_+_49255706 | 0.87 |
ENSDART00000156866
|
si:dkey-183k8.2
|
si:dkey-183k8.2 |
| chr14_-_733565 | 0.86 |
ENSDART00000158097
|
tlr1
|
toll-like receptor 1 |
| chr24_+_5935377 | 0.86 |
ENSDART00000191989
ENSDART00000185932 ENSDART00000131768 |
abi1a
|
abl-interactor 1a |
| chr10_-_22117121 | 0.86 |
ENSDART00000149470
|
si:ch73-111e15.1
|
si:ch73-111e15.1 |
| chr21_-_40835069 | 0.86 |
ENSDART00000004686
|
limk1b
|
LIM domain kinase 1b |
| chr19_-_24745317 | 0.85 |
ENSDART00000142774
|
MYO1G
|
si:dkeyp-92c9.3 |
| chr20_+_53577502 | 0.85 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr13_-_48058779 | 0.85 |
ENSDART00000186623
ENSDART00000045475 |
itga9
|
integrin, alpha 9 |
| chr21_-_37733571 | 0.85 |
ENSDART00000176214
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr20_+_9128256 | 0.84 |
ENSDART00000163883
ENSDART00000183072 ENSDART00000187276 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr2_-_37280617 | 0.84 |
ENSDART00000190458
|
nadkb
|
NAD kinase b |
| chr19_-_3255725 | 0.83 |
ENSDART00000105165
|
illr1
|
immune-related, lectin-like receptor 1 |
| chr1_-_49558223 | 0.83 |
ENSDART00000138510
|
si:ch211-281g13.4
|
si:ch211-281g13.4 |
| chr3_-_5413018 | 0.82 |
ENSDART00000063138
|
vmo1a
|
vitelline membrane outer layer 1 homolog a |
| chr12_+_33484458 | 0.81 |
ENSDART00000000069
|
slc9a3r1a
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1a |
| chr22_+_19453822 | 0.81 |
ENSDART00000141371
|
si:dkey-78l4.6
|
si:dkey-78l4.6 |
| chr17_+_20616576 | 0.78 |
ENSDART00000153561
|
si:ch73-306e8.2
|
si:ch73-306e8.2 |
| chr11_+_14284866 | 0.78 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr3_-_34107685 | 0.77 |
ENSDART00000151130
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr2_-_10877228 | 0.77 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr21_-_14310159 | 0.77 |
ENSDART00000155097
|
si:ch211-196i2.1
|
si:ch211-196i2.1 |
| chr13_-_43238578 | 0.76 |
ENSDART00000161017
|
eif3s6ip
|
eukaryotic translation initiation factor 3, subunit 6 interacting protein |
| chr12_+_6002715 | 0.76 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr6_+_13201358 | 0.76 |
ENSDART00000190290
|
CT009620.1
|
|
| chr6_+_27923054 | 0.75 |
ENSDART00000136833
ENSDART00000145533 |
cep63
|
centrosomal protein 63 |
| chr14_-_46173265 | 0.75 |
ENSDART00000164321
|
zmp:0000000758
|
zmp:0000000758 |
| chr23_+_25354856 | 0.75 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr16_-_22585289 | 0.74 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr8_+_26292560 | 0.74 |
ENSDART00000053463
|
mgll
|
monoglyceride lipase |
| chr8_-_25728628 | 0.74 |
ENSDART00000127237
|
foxp3a
|
forkhead box P3a |
| chr19_+_30867845 | 0.74 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr2_+_11028923 | 0.73 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr24_+_24726956 | 0.73 |
ENSDART00000144574
ENSDART00000066628 |
mtfr1
|
mitochondrial fission regulator 1 |
| chr19_+_40861853 | 0.72 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr5_+_42141917 | 0.72 |
ENSDART00000172201
ENSDART00000140743 |
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr16_+_3185541 | 0.72 |
ENSDART00000024088
|
wdr21
|
WD repeat domain 21 |
| chr7_-_17627320 | 0.71 |
ENSDART00000101701
|
nitr6a
|
novel immune-type receptor 6a |
| chr23_-_4925641 | 0.71 |
ENSDART00000140861
ENSDART00000060718 |
taz
|
tafazzin |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.6 | 2.6 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.5 | 1.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.4 | 2.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.4 | 1.6 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.4 | 1.2 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.4 | 1.2 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.4 | 2.8 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.4 | 1.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.3 | 1.4 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.3 | 1.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.3 | 1.9 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.3 | 2.0 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.3 | 1.7 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.3 | 2.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.3 | 1.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.3 | 1.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 1.4 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 1.6 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.2 | 2.3 | GO:0006706 | steroid catabolic process(GO:0006706) cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 0.6 | GO:0001562 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.2 | 3.0 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.2 | 2.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 0.6 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.2 | 3.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 0.6 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 0.9 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 0.5 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 1.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 1.0 | GO:0031284 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 0.8 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 4.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 2.8 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 0.8 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.4 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 1.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.9 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 1.8 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.4 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 3.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.6 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.6 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 3.9 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 0.9 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.1 | 2.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 1.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 3.1 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 0.4 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 1.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 3.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.6 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 2.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.5 | GO:0006203 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 1.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 1.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.7 | GO:0045823 | positive regulation of heart contraction(GO:0045823) |
| 0.1 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 2.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 1.6 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 0.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 1.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.5 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 1.5 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.0 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.0 | GO:0097696 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 2.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.8 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.7 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 2.3 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 4.8 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.8 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.4 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 1.5 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.0 | 0.9 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.1 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.8 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.7 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 1.3 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 1.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 1.9 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 1.3 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 1.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 2.7 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 1.1 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 1.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 1.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 4.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 1.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.3 | 1.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 0.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 1.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 0.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 0.5 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 3.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.1 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.5 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 3.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 3.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 4.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 7.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 3.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.3 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 2.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.6 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.9 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.8 | GO:0000922 | spindle pole(GO:0000922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.7 | 2.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.5 | 2.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.4 | 2.3 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.3 | 1.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 1.6 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 0.9 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.3 | 1.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 1.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 1.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 1.0 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.2 | 1.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 2.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 1.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 0.6 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.2 | 0.6 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.2 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 4.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 2.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 3.0 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 1.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.7 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.6 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 2.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 2.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.6 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.6 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.5 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.1 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.6 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 2.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.9 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.3 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.1 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 2.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 4.8 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.8 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 3.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 3.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.6 | GO:0046906 | tetrapyrrole binding(GO:0046906) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 5.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 4.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 2.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 2.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.6 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |