Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
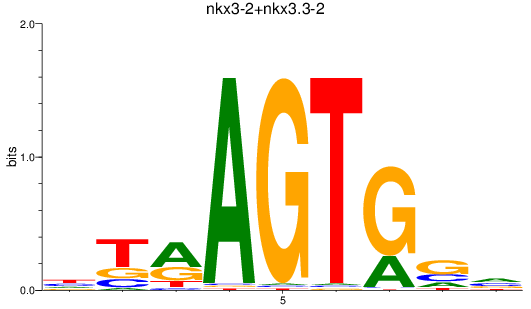
Results for nkx3-2+nkx3.3-2
Z-value: 0.49
Transcription factors associated with nkx3-2+nkx3.3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3-2
|
ENSDARG00000037639 | NK3 homeobox 2 |
|
nkx3.3-2
|
ENSDARG00000069327 | NK3 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx3.3 | dr11_v1_chr13_-_40401870_40401870 | 0.48 | 1.3e-06 | Click! |
| nkx3.2 | dr11_v1_chr14_-_215051_215051 | 0.36 | 3.8e-04 | Click! |
Activity profile of nkx3-2+nkx3.3-2 motif
Sorted Z-values of nkx3-2+nkx3.3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_1789357 | 5.21 |
ENSDART00000006449
|
atp1a1a.2
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 2 |
| chr16_+_26612401 | 3.30 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr17_+_6563307 | 2.49 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr19_+_19989380 | 2.47 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr6_-_40038543 | 2.29 |
ENSDART00000154792
|
si:dkey-197j19.6
|
si:dkey-197j19.6 |
| chr10_+_13209580 | 2.16 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr16_+_49005321 | 2.08 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr18_-_22094102 | 2.01 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr5_-_26765188 | 1.79 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr8_+_24281512 | 1.79 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr4_+_19534833 | 1.72 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr1_-_18803919 | 1.66 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr5_-_26764880 | 1.63 |
ENSDART00000140392
ENSDART00000134728 |
rnf181
|
ring finger protein 181 |
| chr22_+_19188809 | 1.62 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr2_+_42135719 | 1.45 |
ENSDART00000008268
|
cyp7b1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr11_+_14333441 | 1.41 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr15_-_12319065 | 1.33 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr4_+_72668095 | 1.27 |
ENSDART00000182282
ENSDART00000162637 |
CABZ01054391.1
|
|
| chr5_-_54300725 | 1.27 |
ENSDART00000163048
|
slc25a35
|
solute carrier family 25, member 35 |
| chr8_+_25299069 | 1.25 |
ENSDART00000114676
|
gstm.2
|
glutathione S-transferase mu tandem duplicate 2 |
| chr6_-_39198912 | 1.24 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr15_+_15173611 | 1.22 |
ENSDART00000155267
|
si:ch211-149e23.4
|
si:ch211-149e23.4 |
| chr19_-_31802296 | 1.21 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr15_-_36347858 | 1.03 |
ENSDART00000155274
ENSDART00000157936 |
si:dkey-23k10.2
|
si:dkey-23k10.2 |
| chr18_-_29977431 | 1.01 |
ENSDART00000135357
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr9_-_9225980 | 1.01 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr3_-_53559581 | 0.97 |
ENSDART00000183499
|
notch3
|
notch 3 |
| chr2_-_51644044 | 0.97 |
ENSDART00000157899
|
dad1
|
defender against cell death 1 |
| chr15_-_35246742 | 0.96 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr4_-_71436560 | 0.96 |
ENSDART00000166884
|
si:dkey-82i20.1
|
si:dkey-82i20.1 |
| chr7_+_44715224 | 0.94 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr13_+_23093743 | 0.94 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr11_-_36474306 | 0.88 |
ENSDART00000170678
ENSDART00000123591 |
usp48
|
ubiquitin specific peptidase 48 |
| chr12_-_23009312 | 0.87 |
ENSDART00000111801
|
mkxa
|
mohawk homeobox a |
| chr22_+_2830703 | 0.85 |
ENSDART00000145463
ENSDART00000144785 |
si:dkey-20i20.8
|
si:dkey-20i20.8 |
| chr21_-_2348838 | 0.81 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr15_+_888704 | 0.81 |
ENSDART00000182796
|
si:dkey-7i4.9
|
si:dkey-7i4.9 |
| chr12_+_22580579 | 0.79 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr1_-_51710225 | 0.79 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr5_-_42083363 | 0.78 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr10_-_29831944 | 0.77 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr16_+_28994709 | 0.77 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr8_-_17987547 | 0.74 |
ENSDART00000112699
ENSDART00000061747 |
fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr20_+_25568694 | 0.74 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr24_+_21676921 | 0.74 |
ENSDART00000066677
|
si:ch211-140b10.6
|
si:ch211-140b10.6 |
| chr3_-_16413606 | 0.70 |
ENSDART00000127309
ENSDART00000017172 ENSDART00000136465 |
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr1_+_16621345 | 0.70 |
ENSDART00000149026
|
pcm1
|
pericentriolar material 1 |
| chr8_-_48847772 | 0.70 |
ENSDART00000122458
|
wrap73
|
WD repeat containing, antisense to TP73 |
| chr4_+_71018579 | 0.70 |
ENSDART00000186727
|
si:dkeyp-80d11.10
|
si:dkeyp-80d11.10 |
| chr9_-_34882516 | 0.68 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr5_-_31716713 | 0.66 |
ENSDART00000131443
|
dpm2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chr3_-_32603191 | 0.64 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr5_+_26765275 | 0.63 |
ENSDART00000144169
|
si:ch211-102c2.8
|
si:ch211-102c2.8 |
| chr24_+_17005647 | 0.61 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr13_+_15581270 | 0.60 |
ENSDART00000189880
ENSDART00000190067 ENSDART00000041293 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr18_-_38216584 | 0.59 |
ENSDART00000144622
|
si:dkey-10o6.2
|
si:dkey-10o6.2 |
| chr7_-_52842007 | 0.59 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr17_+_31742923 | 0.55 |
ENSDART00000086696
|
arhgap5
|
Rho GTPase activating protein 5 |
| chr3_-_53559408 | 0.53 |
ENSDART00000073930
|
notch3
|
notch 3 |
| chr2_+_39618951 | 0.52 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr3_-_16039619 | 0.51 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr7_+_39688208 | 0.50 |
ENSDART00000189682
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr8_+_25254435 | 0.49 |
ENSDART00000143554
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr2_-_55797318 | 0.49 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr1_-_48933 | 0.49 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr18_-_26781616 | 0.48 |
ENSDART00000136776
ENSDART00000076484 |
kti12
|
KTI12 chromatin associated homolog |
| chr4_+_17327704 | 0.46 |
ENSDART00000016075
ENSDART00000133160 |
nup37
|
nucleoporin 37 |
| chr16_-_13623928 | 0.44 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr8_+_20455134 | 0.43 |
ENSDART00000079618
|
rexo1
|
REX1, RNA exonuclease 1 homolog |
| chr15_-_31027112 | 0.40 |
ENSDART00000100185
|
lgals9l4
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 4 |
| chr4_+_71382288 | 0.39 |
ENSDART00000181926
|
si:ch211-76m11.8
|
si:ch211-76m11.8 |
| chr7_+_20917966 | 0.39 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr4_-_13931508 | 0.38 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr16_-_31933740 | 0.37 |
ENSDART00000125411
|
si:ch1073-90m23.1
|
si:ch1073-90m23.1 |
| chr2_+_42177113 | 0.35 |
ENSDART00000056441
|
tmeff1a
|
transmembrane protein with EGF-like and two follistatin-like domains 1a |
| chr11_-_5953636 | 0.35 |
ENSDART00000140960
ENSDART00000123601 |
dda1
|
DET1 and DDB1 associated 1 |
| chr23_+_12840080 | 0.34 |
ENSDART00000081016
ENSDART00000121697 |
smc1al
|
structural maintenance of chromosomes 1A, like |
| chr21_-_11791909 | 0.34 |
ENSDART00000180893
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr1_-_14258409 | 0.34 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr18_+_22174630 | 0.30 |
ENSDART00000089549
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr14_-_21219659 | 0.29 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr15_+_5360407 | 0.29 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr12_+_17754859 | 0.27 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr21_+_34814444 | 0.26 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr5_-_68779747 | 0.25 |
ENSDART00000192636
ENSDART00000188039 |
mepce
|
methylphosphate capping enzyme |
| chr20_-_3086515 | 0.23 |
ENSDART00000046641
|
map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr24_-_31223232 | 0.23 |
ENSDART00000164155
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr3_-_18384501 | 0.23 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr8_+_28547687 | 0.22 |
ENSDART00000110291
|
srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr2_-_59157790 | 0.22 |
ENSDART00000192303
ENSDART00000159362 |
ftr32
|
finTRIM family, member 32 |
| chr14_-_26392475 | 0.20 |
ENSDART00000170614
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr4_-_75812937 | 0.16 |
ENSDART00000125096
|
si:ch211-203c5.3
|
si:ch211-203c5.3 |
| chr6_-_10168822 | 0.16 |
ENSDART00000151016
|
b3galt1a
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1a |
| chr19_-_3777217 | 0.15 |
ENSDART00000160510
|
si:dkey-206d17.15
|
si:dkey-206d17.15 |
| chr1_+_7988052 | 0.13 |
ENSDART00000167552
|
CR855320.2
|
|
| chr22_+_18389271 | 0.12 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr1_-_28860732 | 0.11 |
ENSDART00000177588
|
timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr12_+_13404784 | 0.11 |
ENSDART00000167977
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr11_-_1948784 | 0.11 |
ENSDART00000082475
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr25_+_19670273 | 0.10 |
ENSDART00000073472
|
zgc:113426
|
zgc:113426 |
| chr22_+_9431853 | 0.09 |
ENSDART00000181028
|
si:ch211-11p18.6
|
si:ch211-11p18.6 |
| chr16_-_16522013 | 0.08 |
ENSDART00000160602
|
nbeal2
|
neurobeachin-like 2 |
| chr1_-_28861226 | 0.06 |
ENSDART00000075502
|
timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr21_-_38031038 | 0.04 |
ENSDART00000179483
ENSDART00000076238 |
rbm41
|
RNA binding motif protein 41 |
| chr25_+_16356083 | 0.02 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr22_+_9922301 | 0.00 |
ENSDART00000105924
|
blf
|
bloody fingers |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3-2+nkx3.3-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0002544 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.5 | 1.5 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.3 | 1.0 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.3 | 5.2 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 1.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.2 | 2.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.2 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.4 | GO:2000561 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.4 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.9 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.7 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 2.0 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 1.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 1.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.6 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.8 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.1 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.7 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.7 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 1.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 2.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.7 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.3 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 0.7 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.2 | 1.0 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 1.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.8 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 2.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 2.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 2.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0017069 | snRNA binding(GO:0017069) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 1.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 1.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 1.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |