Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
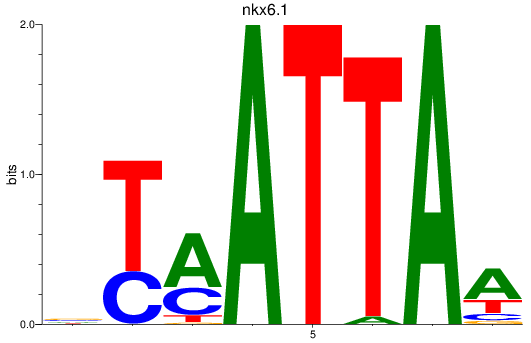
Results for nkx6.1
Z-value: 1.00
Transcription factors associated with nkx6.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx6.1
|
ENSDARG00000022569 | NK6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx6.1 | dr11_v1_chr21_+_19070921_19070921 | 0.58 | 8.7e-10 | Click! |
Activity profile of nkx6.1 motif
Sorted Z-values of nkx6.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_32753535 | 12.25 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr17_+_15433518 | 12.22 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433671 | 12.19 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr16_+_46111849 | 11.26 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr17_+_23298928 | 8.14 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr9_+_31282161 | 7.39 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr16_+_5774977 | 7.24 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr14_+_50770537 | 7.18 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr7_+_25059845 | 7.16 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr21_+_6780340 | 7.11 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr13_+_38430466 | 6.88 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr12_+_24342303 | 6.69 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr10_-_24371312 | 6.46 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr10_+_29698467 | 6.45 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr17_-_12389259 | 6.32 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr16_+_20161805 | 6.30 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr14_+_14662116 | 6.26 |
ENSDART00000161693
|
cetn2
|
centrin, EF-hand protein, 2 |
| chr17_-_16965809 | 6.18 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr15_-_16098531 | 6.16 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr19_-_5103313 | 6.08 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr23_+_40460333 | 6.04 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr20_+_18551657 | 6.00 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr9_-_31278048 | 6.00 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr18_+_9637744 | 5.95 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr23_+_28731379 | 5.93 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr7_-_41014773 | 5.81 |
ENSDART00000013785
|
insig1
|
insulin induced gene 1 |
| chr18_-_1185772 | 5.73 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr4_+_21129752 | 5.63 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr19_-_5103141 | 5.46 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr5_-_23362602 | 5.45 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr21_+_13366353 | 5.44 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr6_-_41229787 | 5.35 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr24_-_29963858 | 5.34 |
ENSDART00000183442
|
CR352310.1
|
|
| chr22_-_24738188 | 5.34 |
ENSDART00000050238
|
vtg1
|
vitellogenin 1 |
| chr1_+_12766351 | 5.33 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr5_-_25723079 | 5.31 |
ENSDART00000014013
|
gda
|
guanine deaminase |
| chr1_-_50859053 | 5.22 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr4_+_12615836 | 5.22 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr5_-_50992690 | 5.15 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr8_+_24861264 | 5.13 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr24_+_2519761 | 5.07 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr16_+_23913943 | 5.05 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr5_-_31926906 | 4.99 |
ENSDART00000187340
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr23_+_20563779 | 4.97 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr7_+_19552381 | 4.88 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr13_+_25720969 | 4.73 |
ENSDART00000046050
|
pcbd1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr20_-_28800999 | 4.72 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr6_-_10320676 | 4.69 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr12_-_28881638 | 4.68 |
ENSDART00000148459
ENSDART00000039667 ENSDART00000148668 ENSDART00000136593 ENSDART00000139923 ENSDART00000148912 |
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr5_+_20147830 | 4.67 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr22_-_13851297 | 4.66 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr8_-_25120231 | 4.62 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr22_-_24791505 | 4.60 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr15_+_22311803 | 4.55 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr8_-_34052019 | 4.54 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr12_+_31713239 | 4.49 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr7_-_28148310 | 4.47 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr11_-_37509001 | 4.46 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr2_-_55298075 | 4.43 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr21_-_39177564 | 4.42 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr20_-_9436521 | 4.40 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr2_+_20332044 | 4.38 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr2_+_50608099 | 4.37 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr19_-_31522625 | 4.35 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr21_+_27416284 | 4.22 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr7_-_69521481 | 4.16 |
ENSDART00000148465
|
slc1a1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr11_+_30057762 | 4.13 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr19_-_32641725 | 3.99 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr21_-_42100471 | 3.96 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr3_+_33341640 | 3.93 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr3_+_29714775 | 3.88 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr21_+_28958471 | 3.88 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr25_-_13842618 | 3.88 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr16_-_42965192 | 3.87 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr18_+_34478959 | 3.87 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr13_+_38521152 | 3.86 |
ENSDART00000145292
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr13_+_23988442 | 3.81 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr1_-_45042210 | 3.78 |
ENSDART00000073694
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr25_+_15273370 | 3.78 |
ENSDART00000045659
|
tcp11l1
|
t-complex 11, testis-specific-like 1 |
| chr5_+_63668735 | 3.76 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr5_+_19314574 | 3.74 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr12_+_18681477 | 3.62 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr19_+_6938289 | 3.60 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr11_+_30244356 | 3.58 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr14_-_1454045 | 3.57 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr3_-_18711288 | 3.56 |
ENSDART00000183885
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr23_-_26522760 | 3.55 |
ENSDART00000142417
ENSDART00000135606 ENSDART00000122668 |
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr24_-_38110779 | 3.53 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr3_-_13146631 | 3.53 |
ENSDART00000172460
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr11_+_13223625 | 3.53 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr9_-_20372977 | 3.52 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr9_+_38163876 | 3.50 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr9_-_3671911 | 3.49 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr14_+_49135264 | 3.45 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr12_+_5081759 | 3.45 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr1_+_8601935 | 3.45 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr13_+_36622100 | 3.41 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr11_-_27501027 | 3.40 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr6_-_44044385 | 3.38 |
ENSDART00000075497
|
rybpb
|
RING1 and YY1 binding protein b |
| chr2_-_5475910 | 3.38 |
ENSDART00000100954
ENSDART00000172143 ENSDART00000132496 |
proca
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr24_-_21923930 | 3.37 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr5_-_50781623 | 3.34 |
ENSDART00000114950
|
zgc:194908
|
zgc:194908 |
| chr19_-_3931917 | 3.33 |
ENSDART00000162532
|
map7d1b
|
MAP7 domain containing 1b |
| chr10_+_22381802 | 3.32 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr21_-_35325466 | 3.30 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr16_-_28658341 | 3.29 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr1_+_33969015 | 3.28 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr20_+_40457599 | 3.24 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr20_+_31076488 | 3.23 |
ENSDART00000136255
ENSDART00000008840 |
otofa
|
otoferlin a |
| chr22_-_12160283 | 3.22 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr19_-_31402429 | 3.22 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr4_-_8903240 | 3.20 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr10_+_37137464 | 3.16 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr7_-_24699985 | 3.13 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr10_+_7593185 | 3.11 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr1_-_44704261 | 3.11 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr14_-_4273396 | 3.08 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr11_-_8126223 | 3.06 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr4_+_5798223 | 3.05 |
ENSDART00000059440
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr20_-_9462433 | 3.04 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr1_+_40023640 | 3.04 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr12_-_33972798 | 3.02 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr22_-_24818066 | 3.02 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr3_+_28860283 | 3.01 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr18_-_18875308 | 3.01 |
ENSDART00000127182
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr14_-_1454413 | 2.99 |
ENSDART00000185403
ENSDART00000191357 |
pmt
|
phosphoethanolamine methyltransferase |
| chr13_-_30700460 | 2.99 |
ENSDART00000139073
|
rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr18_+_10884996 | 2.98 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr2_-_5466708 | 2.97 |
ENSDART00000136682
|
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr22_-_20166660 | 2.95 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr18_-_2433011 | 2.94 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr9_+_24159280 | 2.93 |
ENSDART00000184624
ENSDART00000178422 |
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr10_-_34871737 | 2.92 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr8_-_50888806 | 2.92 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr3_+_34919810 | 2.91 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr15_+_8767650 | 2.87 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr11_+_23760470 | 2.87 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr16_-_17162843 | 2.86 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr1_-_44701313 | 2.86 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr12_-_35787801 | 2.83 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr2_+_38147761 | 2.83 |
ENSDART00000135307
|
sall2
|
spalt-like transcription factor 2 |
| chr15_-_20024205 | 2.81 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr11_+_36158134 | 2.80 |
ENSDART00000189827
ENSDART00000163330 |
grm2b
|
glutamate receptor, metabotropic 2b |
| chr18_+_24921587 | 2.79 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr16_+_17389116 | 2.77 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr19_-_7358184 | 2.75 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr10_+_21867307 | 2.74 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr19_+_21362553 | 2.74 |
ENSDART00000122002
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr16_+_34531486 | 2.74 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr7_-_28147838 | 2.73 |
ENSDART00000158921
|
lmo1
|
LIM domain only 1 |
| chr1_+_34181581 | 2.73 |
ENSDART00000146042
|
epha6
|
eph receptor A6 |
| chr6_+_21001264 | 2.71 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr5_+_36611128 | 2.68 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr11_-_19775182 | 2.67 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr14_-_8271686 | 2.67 |
ENSDART00000165120
|
purab
|
purine-rich element binding protein Ab |
| chr6_-_38419318 | 2.67 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr24_+_5208171 | 2.67 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr23_+_28648864 | 2.65 |
ENSDART00000189096
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr11_-_6188413 | 2.61 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr8_+_7144066 | 2.61 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr13_-_31435137 | 2.61 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr23_+_37955041 | 2.59 |
ENSDART00000087148
|
cbln4
|
cerebellin 4 precursor |
| chr1_+_25801648 | 2.59 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr7_+_71547747 | 2.59 |
ENSDART00000180869
|
adcyap1a
|
adenylate cyclase activating polypeptide 1a |
| chr6_-_35779348 | 2.59 |
ENSDART00000191159
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr8_-_30979494 | 2.55 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr15_-_6247775 | 2.53 |
ENSDART00000148350
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr23_+_30730121 | 2.53 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr16_+_24681177 | 2.52 |
ENSDART00000058956
ENSDART00000189335 |
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr21_-_5856050 | 2.52 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr20_-_40755614 | 2.51 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr21_+_15870752 | 2.50 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr13_-_10945288 | 2.48 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr20_+_27087539 | 2.47 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr22_+_5176255 | 2.47 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr7_+_25033924 | 2.47 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr20_+_41756996 | 2.46 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr3_-_20040636 | 2.43 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr3_-_28258462 | 2.42 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr20_-_35040041 | 2.42 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr2_-_5135125 | 2.42 |
ENSDART00000164039
|
ptmab
|
prothymosin, alpha b |
| chr1_-_22512063 | 2.41 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr23_-_38497705 | 2.39 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr3_-_35800221 | 2.39 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr16_+_28578352 | 2.38 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr9_-_1702648 | 2.37 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr20_-_20931197 | 2.36 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr15_-_16177603 | 2.35 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr11_-_12998400 | 2.35 |
ENSDART00000018614
|
chrna4b
|
cholinergic receptor, nicotinic, alpha 4b |
| chr15_-_34213898 | 2.31 |
ENSDART00000191945
ENSDART00000186089 |
etv1
|
ets variant 1 |
| chr22_-_15010688 | 2.30 |
ENSDART00000139892
|
elfn2a
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2a |
| chr20_-_19511700 | 2.29 |
ENSDART00000040191
|
snx17
|
sorting nexin 17 |
| chr10_-_43029001 | 2.29 |
ENSDART00000171494
|
ssbp2
|
single-stranded DNA binding protein 2 |
| chr16_+_25011994 | 2.28 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr21_-_25604603 | 2.28 |
ENSDART00000133134
ENSDART00000139460 |
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr17_-_31611692 | 2.28 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr1_+_11977426 | 2.28 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr14_-_24110707 | 2.27 |
ENSDART00000133522
ENSDART00000123152 |
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr23_+_20705849 | 2.26 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr4_-_17629444 | 2.25 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx6.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 2.9 | 11.5 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 2.1 | 10.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 1.9 | 5.8 | GO:0036315 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.9 | 13.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 1.8 | 5.3 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 1.7 | 5.1 | GO:0015824 | proline transport(GO:0015824) |
| 1.6 | 4.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 1.2 | 12.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 1.2 | 3.6 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 1.1 | 4.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 1.0 | 3.9 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.9 | 3.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.8 | 4.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.8 | 3.1 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.7 | 0.7 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.6 | 2.6 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.6 | 4.3 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.6 | 5.0 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.6 | 1.8 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.6 | 4.7 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.6 | 2.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.6 | 4.4 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.5 | 1.6 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.5 | 1.6 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.5 | 1.4 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.5 | 4.7 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.5 | 2.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.5 | 6.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 2.2 | GO:0071543 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.4 | 2.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 1.3 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.4 | 1.3 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 2.2 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.4 | 1.8 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.4 | 2.9 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.3 | 13.5 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.3 | 6.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 1.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 1.7 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.3 | 1.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.3 | 6.7 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.3 | 7.2 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.3 | 2.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 3.3 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.3 | 3.0 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.3 | 1.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.3 | 6.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 1.5 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.3 | 8.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.3 | 1.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.3 | 3.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.3 | 2.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.3 | 5.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 3.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.2 | 2.0 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.2 | 1.0 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.2 | 1.2 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.2 | 0.7 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 2.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 0.7 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.2 | 3.4 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.2 | 3.8 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.2 | 1.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.2 | 5.3 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.2 | 3.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.0 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.2 | 4.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.6 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.2 | 2.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 2.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.2 | 3.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 0.8 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.2 | 0.7 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.2 | 4.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 5.2 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.2 | 0.5 | GO:0097065 | anterior head development(GO:0097065) |
| 0.2 | 0.7 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.2 | 7.5 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 4.3 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.2 | 0.5 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 1.2 | GO:0042311 | vasodilation(GO:0042311) |
| 0.2 | 1.8 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.2 | 3.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 0.6 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.2 | 2.7 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 0.5 | GO:0034380 | plasma lipoprotein particle assembly(GO:0034377) high-density lipoprotein particle assembly(GO:0034380) chylomicron remnant clearance(GO:0034382) protein-lipid complex assembly(GO:0065005) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.2 | 0.2 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.2 | 1.2 | GO:0030643 | microglia differentiation(GO:0014004) cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 2.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 2.7 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 4.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 0.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 4.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 1.0 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.3 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 2.7 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 0.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 2.0 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 1.4 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 1.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 2.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 2.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 3.5 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.1 | 6.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.5 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.7 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 5.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.8 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.1 | 1.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 1.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 7.6 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 4.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 2.2 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 5.9 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.1 | 1.2 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 0.6 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 0.8 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 1.5 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 2.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 1.0 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.0 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 2.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 1.4 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 10.6 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 1.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 6.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 1.2 | GO:0045116 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 1.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 2.9 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 2.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 3.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.2 | GO:0050805 | negative regulation of synaptic transmission(GO:0050805) |
| 0.1 | 0.7 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 2.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.5 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 1.2 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.1 | 0.3 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.1 | 9.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 1.5 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 0.6 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 1.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 2.6 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 0.7 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) synaptic membrane adhesion(GO:0099560) |
| 0.0 | 7.0 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 2.3 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 2.4 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 4.2 | GO:0006956 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 1.4 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 0.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 2.9 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.4 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.8 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.5 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 1.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 1.2 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.9 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 1.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 2.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 6.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 2.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 3.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.5 | GO:0008406 | gonad development(GO:0008406) |
| 0.0 | 0.2 | GO:1900117 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 1.0 | GO:0072666 | protein targeting to vacuole(GO:0006623) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 1.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.4 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.5 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 5.8 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.1 | GO:0060898 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 1.1 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 1.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 5.4 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 4.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.4 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 4.4 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 1.6 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.7 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.7 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.1 | GO:0034627 | tryptophan catabolic process to kynurenine(GO:0019441) 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.4 | GO:0010675 | regulation of carbohydrate metabolic process(GO:0006109) regulation of cellular carbohydrate metabolic process(GO:0010675) |
| 0.0 | 0.5 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 0.1 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.1 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.8 | 10.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.2 | 12.4 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 1.2 | 3.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 1.1 | 5.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.1 | 3.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.1 | 6.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.9 | 3.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.6 | 3.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.6 | 1.7 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.5 | 2.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.5 | 3.9 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.5 | 4.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.4 | 4.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.4 | 3.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 2.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.4 | 3.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 10.3 | GO:0044306 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.3 | 6.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 2.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 2.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 23.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 0.9 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 1.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 8.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 3.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 9.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 6.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 4.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 0.8 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.2 | 2.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 4.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 1.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 0.7 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 10.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 2.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.8 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.0 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.7 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 0.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 2.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.6 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 4.1 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 2.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.8 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 15.4 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.4 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 4.4 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 2.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 6.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 5.6 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 3.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 3.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 7.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 11.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.7 | GO:0035770 | ribonucleoprotein granule(GO:0035770) |
| 0.0 | 10.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 3.3 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.5 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 2.4 | 7.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.9 | 13.0 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 1.6 | 24.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 1.5 | 4.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 1.2 | 4.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.0 | 5.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.9 | 6.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.9 | 3.5 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.8 | 3.9 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.8 | 6.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.7 | 12.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.6 | 3.9 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.6 | 2.6 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.6 | 1.7 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.5 | 2.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.5 | 2.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.5 | 2.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 2.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.5 | 3.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 3.0 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.5 | 6.6 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.5 | 7.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.5 | 1.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.4 | 2.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.4 | 2.2 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.4 | 3.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.4 | 4.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.4 | 2.8 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.4 | 1.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.4 | 3.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 1.9 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.4 | 2.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 1.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.4 | 6.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 5.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.4 | 6.7 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.4 | 1.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.3 | 1.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.3 | 11.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 1.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 2.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 2.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 3.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 2.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.3 | 1.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 8.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 2.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 3.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 1.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 2.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 3.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 4.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 1.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 4.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 1.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 0.7 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.2 | 2.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 4.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 7.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 1.4 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.2 | 0.6 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.2 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 3.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 3.8 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 1.0 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 2.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 2.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 3.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 5.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 2.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 2.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 3.1 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 1.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 2.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 2.0 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.8 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 5.2 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.4 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 4.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 3.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 3.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 5.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 10.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 4.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 2.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 2.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.5 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 3.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 6.0 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 9.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 4.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.5 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 1.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 9.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 4.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 2.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 3.0 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 8.6 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 5.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 7.5 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 3.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 6.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 3.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 6.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 2.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.1 | 4.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.9 | 11.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.7 | 10.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.7 | 1.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.6 | 4.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.6 | 7.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.5 | 4.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.5 | 6.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 3.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 2.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 2.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 1.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 6.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 7.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 3.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.8 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 3.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |