Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
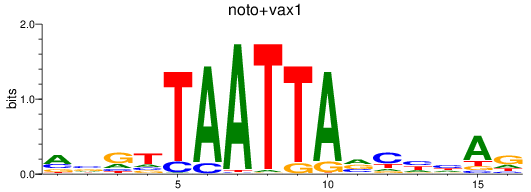
Results for noto+vax1
Z-value: 0.45
Transcription factors associated with noto+vax1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
noto
|
ENSDARG00000021201 | notochord homeobox |
|
vax1
|
ENSDARG00000021916 | ventral anterior homeobox 1 |
|
vax1
|
ENSDARG00000114230 | ventral anterior homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| noto | dr11_v1_chr13_+_14976108_14976108 | 0.17 | 1.1e-01 | Click! |
| vax1 | dr11_v1_chr17_-_21441464_21441464 | -0.11 | 2.9e-01 | Click! |
Activity profile of noto+vax1 motif
Sorted Z-values of noto+vax1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_55982300 | 3.29 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr19_+_2631565 | 2.88 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr13_-_35808904 | 2.62 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr15_-_43284021 | 2.51 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr9_+_38372216 | 2.50 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr4_+_73085993 | 2.45 |
ENSDART00000165749
|
si:ch73-170d6.2
|
si:ch73-170d6.2 |
| chr21_-_35419486 | 2.35 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr8_+_22516728 | 2.27 |
ENSDART00000146013
|
si:ch211-261n11.3
|
si:ch211-261n11.3 |
| chr14_+_51056605 | 2.25 |
ENSDART00000159639
|
CABZ01078593.1
|
|
| chr3_+_59784632 | 2.24 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr12_+_22580579 | 2.21 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr17_+_16046314 | 2.14 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr3_+_27798094 | 1.97 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr19_-_2861444 | 1.95 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr24_+_19415124 | 1.89 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr3_-_15734530 | 1.83 |
ENSDART00000141142
|
mvp
|
major vault protein |
| chr20_+_26966725 | 1.77 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr3_-_15734358 | 1.75 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr6_-_40581376 | 1.68 |
ENSDART00000185412
|
tspo
|
translocator protein |
| chr10_-_33621739 | 1.64 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr9_+_30421489 | 1.63 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr22_-_30770751 | 1.52 |
ENSDART00000172115
|
AL831726.2
|
|
| chr7_-_30174882 | 1.41 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr19_-_7690975 | 1.34 |
ENSDART00000151384
|
si:dkey-204a24.10
|
si:dkey-204a24.10 |
| chr18_-_46256560 | 1.30 |
ENSDART00000171375
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr20_-_37813863 | 1.27 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr6_+_6924637 | 1.27 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr15_+_31701716 | 1.27 |
ENSDART00000113370
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr15_+_31899312 | 1.24 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
| chr9_-_5263947 | 1.21 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr25_+_28825657 | 1.19 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr16_+_13818743 | 1.18 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr19_+_41464870 | 1.18 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr1_-_51719110 | 1.10 |
ENSDART00000190574
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr16_+_13818500 | 1.08 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr1_+_513986 | 1.06 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr14_-_14640401 | 1.04 |
ENSDART00000168027
ENSDART00000167521 |
znf185
|
zinc finger protein 185 with LIM domain |
| chr21_-_26490186 | 1.00 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr4_+_21129752 | 0.93 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr20_-_14925281 | 0.93 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr18_-_2727764 | 0.92 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr22_+_19218733 | 0.86 |
ENSDART00000183212
ENSDART00000133595 |
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr6_-_19271210 | 0.82 |
ENSDART00000163628
ENSDART00000159124 |
zgc:174863
|
zgc:174863 |
| chr15_+_5088210 | 0.81 |
ENSDART00000183423
|
mxf
|
myxovirus (influenza virus) resistance F |
| chr9_-_14683574 | 0.76 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr6_-_30954268 | 0.74 |
ENSDART00000154523
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr4_+_9011448 | 0.73 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr3_-_32902138 | 0.66 |
ENSDART00000144026
ENSDART00000083874 ENSDART00000145443 ENSDART00000148239 ENSDART00000134917 |
kat7a
|
K(lysine) acetyltransferase 7a |
| chr1_-_513762 | 0.66 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr11_+_43043171 | 0.64 |
ENSDART00000180344
|
CABZ01092982.1
|
|
| chr3_-_34084387 | 0.63 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr5_+_41477954 | 0.62 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr7_-_46777876 | 0.61 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr6_+_32382743 | 0.59 |
ENSDART00000190009
|
dock7
|
dedicator of cytokinesis 7 |
| chr17_+_21902058 | 0.58 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr18_+_17827149 | 0.57 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr20_-_46362606 | 0.54 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr3_-_27601812 | 0.53 |
ENSDART00000077734
|
zgc:66160
|
zgc:66160 |
| chr13_+_35339182 | 0.53 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr20_-_14924858 | 0.51 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr11_-_13107106 | 0.49 |
ENSDART00000184477
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr8_+_25034544 | 0.47 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr18_+_3332999 | 0.47 |
ENSDART00000160857
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr22_-_2959005 | 0.46 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr5_+_41477526 | 0.44 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr12_+_20352400 | 0.43 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr2_-_2957970 | 0.41 |
ENSDART00000162505
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr14_+_30762131 | 0.40 |
ENSDART00000145039
|
si:ch211-145o7.3
|
si:ch211-145o7.3 |
| chr2_-_26720854 | 0.40 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr12_+_30265802 | 0.37 |
ENSDART00000182412
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr20_-_1635922 | 0.35 |
ENSDART00000181502
|
CR846082.1
|
|
| chr14_-_4145594 | 0.35 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr13_+_35637048 | 0.31 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr15_+_1796313 | 0.31 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr10_+_43039947 | 0.30 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr20_+_29209615 | 0.29 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr14_+_46313396 | 0.28 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr3_-_27601544 | 0.28 |
ENSDART00000188201
|
zgc:66160
|
zgc:66160 |
| chr20_+_11731039 | 0.26 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr4_-_45100253 | 0.25 |
ENSDART00000163870
|
si:dkey-51d8.3
|
si:dkey-51d8.3 |
| chr17_+_24722646 | 0.24 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr11_+_11303458 | 0.18 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr18_-_43884044 | 0.16 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr14_+_46313135 | 0.14 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr1_+_21731382 | 0.14 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr16_+_11834516 | 0.13 |
ENSDART00000146611
|
cxcr3.3
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 3 |
| chr5_+_48683447 | 0.11 |
ENSDART00000008043
|
adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr19_-_20430892 | 0.11 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr6_+_41191482 | 0.08 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr1_-_7970653 | 0.08 |
ENSDART00000188909
|
si:dkey-79f11.7
|
si:dkey-79f11.7 |
| chr6_+_3280939 | 0.07 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr3_-_50443607 | 0.07 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr5_-_66397688 | 0.07 |
ENSDART00000161483
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr20_+_29209767 | 0.05 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr8_-_52562672 | 0.04 |
ENSDART00000159333
ENSDART00000159974 |
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr5_-_15283509 | 0.02 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr4_+_306036 | 0.02 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr15_-_22074315 | 0.01 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of noto+vax1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 0.7 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 1.9 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 0.8 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 1.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 1.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.7 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.4 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 2.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 2.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 3.6 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.1 | 1.2 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 2.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 2.0 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 2.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 2.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.5 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 2.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 1.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.9 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 2.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.7 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 2.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 2.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.2 | 0.7 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 1.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 1.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 2.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 1.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 4.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |