Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
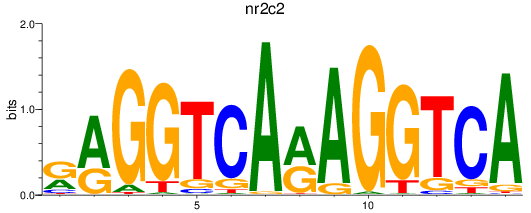
Results for nr2c2
Z-value: 0.77
Transcription factors associated with nr2c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2c2
|
ENSDARG00000042477 | nuclear receptor subfamily 2, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2c2 | dr11_v1_chr8_+_53120278_53120278 | -0.33 | 1.3e-03 | Click! |
Activity profile of nr2c2 motif
Sorted Z-values of nr2c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_36837846 | 19.08 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr3_+_39566999 | 14.76 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr2_+_42191592 | 11.96 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr18_+_1615 | 10.82 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr20_+_15015557 | 10.76 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr11_+_11201096 | 10.46 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr21_-_19006631 | 10.15 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr9_-_22834860 | 9.86 |
ENSDART00000146486
|
neb
|
nebulin |
| chr3_+_30257582 | 9.03 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr22_-_7050 | 8.75 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr7_+_31871830 | 8.40 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr12_-_4683325 | 7.86 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr11_+_77526 | 7.34 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr3_-_12187245 | 7.08 |
ENSDART00000189553
ENSDART00000165131 |
srl
|
sarcalumenin |
| chr4_-_858434 | 6.95 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr6_-_40722480 | 6.94 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr7_+_31838320 | 6.84 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr6_-_40722200 | 6.30 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr16_-_24612871 | 5.95 |
ENSDART00000155614
ENSDART00000154787 ENSDART00000155983 ENSDART00000156519 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr24_-_33703504 | 5.12 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr1_+_58353661 | 4.89 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr10_+_43994471 | 4.53 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr19_-_9648542 | 4.47 |
ENSDART00000172628
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr15_-_29598679 | 4.31 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr6_+_27339962 | 4.11 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr18_-_48508585 | 4.11 |
ENSDART00000133364
|
kcnj1a.4
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 4 |
| chr21_-_35832548 | 4.01 |
ENSDART00000180840
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr16_-_19568388 | 3.99 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr1_-_58868306 | 3.89 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr2_+_57801960 | 3.81 |
ENSDART00000147966
|
si:dkeyp-68b7.10
|
si:dkeyp-68b7.10 |
| chr18_-_48517040 | 3.78 |
ENSDART00000143645
|
kcnj1a.3
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 3 |
| chr21_+_15704556 | 3.74 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr18_-_48492951 | 3.55 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr7_+_22718251 | 3.50 |
ENSDART00000027718
ENSDART00000143341 |
fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr8_+_44549290 | 3.46 |
ENSDART00000160801
|
si:dkey-220o5.5
|
si:dkey-220o5.5 |
| chr10_+_35417099 | 3.44 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr23_+_2740741 | 3.43 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr1_+_58182400 | 3.42 |
ENSDART00000144784
|
si:ch211-15j1.1
|
si:ch211-15j1.1 |
| chr3_-_34561624 | 3.35 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr16_-_24605969 | 3.28 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr1_-_59232267 | 3.21 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr16_-_25233515 | 3.20 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr16_-_22713152 | 3.10 |
ENSDART00000140953
ENSDART00000143836 |
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr18_-_48550426 | 3.01 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr6_+_60036767 | 2.98 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr3_+_58167288 | 2.87 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr24_+_81527 | 2.80 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr17_+_53424415 | 2.79 |
ENSDART00000157022
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr17_+_15788100 | 2.73 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr9_+_54644626 | 2.66 |
ENSDART00000190609
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr14_+_97017 | 2.62 |
ENSDART00000159300
ENSDART00000169523 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr21_-_1640547 | 2.60 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr3_-_32590164 | 2.56 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr12_-_47782623 | 2.56 |
ENSDART00000115742
|
selenou1b
|
selenoprotein U1b |
| chr14_+_15331486 | 2.53 |
ENSDART00000172077
ENSDART00000183370 ENSDART00000182467 |
SPINK2
si:dkey-203a12.5
|
si:dkey-203a12.4 si:dkey-203a12.5 |
| chr3_-_18410968 | 2.52 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr21_+_33454147 | 2.44 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr9_-_43082945 | 2.39 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr5_+_35786141 | 2.39 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr13_-_37254777 | 2.37 |
ENSDART00000139734
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr12_+_46708920 | 2.37 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr4_-_16451375 | 2.36 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr7_+_34297271 | 2.34 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr21_-_308852 | 2.34 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr23_+_42810055 | 2.29 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr19_+_2631565 | 2.29 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr3_+_301479 | 2.25 |
ENSDART00000165169
|
CABZ01075611.1
|
|
| chr19_-_27564458 | 2.24 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr20_+_1996202 | 2.22 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr13_-_35808904 | 2.20 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr16_-_5115993 | 2.19 |
ENSDART00000138654
|
ttk
|
ttk protein kinase |
| chr17_-_8976307 | 2.18 |
ENSDART00000092113
|
zranb1b
|
zinc finger, RAN-binding domain containing 1b |
| chr5_+_22579975 | 2.14 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr1_+_15062 | 2.12 |
ENSDART00000169005
|
cep97
|
centrosomal protein 97 |
| chr12_+_49125510 | 2.11 |
ENSDART00000185804
|
FO704607.1
|
|
| chr7_+_24115082 | 2.11 |
ENSDART00000182718
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr22_-_15587360 | 2.10 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr13_+_28819768 | 2.08 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr8_+_31872992 | 2.08 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr4_-_16833518 | 2.07 |
ENSDART00000179867
|
ldhba
|
lactate dehydrogenase Ba |
| chr23_+_17865554 | 2.05 |
ENSDART00000181009
ENSDART00000162822 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr11_-_6974022 | 2.01 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr14_+_30413758 | 2.01 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr18_+_27926839 | 1.99 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr7_-_2090594 | 1.99 |
ENSDART00000183166
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr20_-_47953524 | 1.98 |
ENSDART00000167986
|
HADHB
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr22_-_24992532 | 1.95 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr10_-_322769 | 1.94 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr23_-_16737161 | 1.89 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr7_+_24114694 | 1.89 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr22_-_28668442 | 1.87 |
ENSDART00000182377
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr8_-_50133064 | 1.87 |
ENSDART00000030064
|
antxr1a
|
anthrax toxin receptor 1a |
| chr3_+_1179601 | 1.86 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr1_+_58312187 | 1.82 |
ENSDART00000142285
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr20_-_49117438 | 1.80 |
ENSDART00000057700
|
naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr16_+_7985886 | 1.79 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr1_+_45072778 | 1.78 |
ENSDART00000172529
|
btr01
|
bloodthirsty-related gene family, member 1 |
| chr5_+_24282570 | 1.74 |
ENSDART00000041905
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr17_+_33418475 | 1.74 |
ENSDART00000169145
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr16_-_19568795 | 1.68 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr5_-_24124118 | 1.66 |
ENSDART00000051550
|
capga
|
capping protein (actin filament), gelsolin-like a |
| chr1_-_44314 | 1.66 |
ENSDART00000160050
|
tmem39a
|
transmembrane protein 39A |
| chr6_-_49873020 | 1.66 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr12_-_979789 | 1.63 |
ENSDART00000128188
|
daglb
|
diacylglycerol lipase, beta |
| chr1_-_52292235 | 1.60 |
ENSDART00000132638
|
si:dkey-121b10.7
|
si:dkey-121b10.7 |
| chr17_+_53425037 | 1.59 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr6_+_52931841 | 1.59 |
ENSDART00000174358
|
si:dkeyp-3f10.12
|
si:dkeyp-3f10.12 |
| chr6_+_1787160 | 1.58 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr25_+_418932 | 1.57 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr7_+_24520518 | 1.55 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr8_+_2530065 | 1.54 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr1_+_58303892 | 1.53 |
ENSDART00000147678
|
CR769768.1
|
|
| chr1_+_58139102 | 1.53 |
ENSDART00000134826
|
si:ch211-15j1.4
|
si:ch211-15j1.4 |
| chr17_-_22137199 | 1.52 |
ENSDART00000089689
|
mrps5
|
mitochondrial ribosomal protein S5 |
| chr3_-_36419641 | 1.51 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr3_+_27770110 | 1.50 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr1_+_57145072 | 1.49 |
ENSDART00000152776
|
si:ch73-94k4.4
|
si:ch73-94k4.4 |
| chr25_+_31238606 | 1.48 |
ENSDART00000149439
|
tnni2a.2
|
troponin I type 2a (skeletal, fast), tandem duplicate 2 |
| chr23_-_45504991 | 1.46 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr14_+_989733 | 1.45 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr23_+_8797143 | 1.44 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr10_-_21362071 | 1.44 |
ENSDART00000125167
|
avd
|
avidin |
| chr1_-_44933094 | 1.42 |
ENSDART00000147527
|
si:dkey-9i23.14
|
si:dkey-9i23.14 |
| chr16_-_34258931 | 1.41 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr1_-_51720633 | 1.40 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr21_-_9446747 | 1.38 |
ENSDART00000158790
|
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr3_-_18805225 | 1.38 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr25_+_20119466 | 1.38 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr16_-_54357200 | 1.38 |
ENSDART00000170753
|
CABZ01111454.1
|
|
| chr8_+_50742975 | 1.38 |
ENSDART00000155664
ENSDART00000160612 |
si:ch73-6l19.2
|
si:ch73-6l19.2 |
| chr7_+_34296789 | 1.38 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr14_+_34501245 | 1.36 |
ENSDART00000131424
|
lcp2b
|
lymphocyte cytosolic protein 2b |
| chr18_+_13315739 | 1.36 |
ENSDART00000143404
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr18_-_43866526 | 1.35 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr23_-_45705525 | 1.34 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr1_+_58119117 | 1.34 |
ENSDART00000146788
|
si:ch211-15j1.5
|
si:ch211-15j1.5 |
| chr8_+_15251448 | 1.34 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr20_+_47953047 | 1.32 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr14_+_740032 | 1.32 |
ENSDART00000165014
|
klb
|
klotho beta |
| chr7_+_59649746 | 1.31 |
ENSDART00000181067
|
rpl34
|
ribosomal protein L34 |
| chr9_+_33216945 | 1.31 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr8_-_979735 | 1.30 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr1_+_58312680 | 1.29 |
ENSDART00000138007
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr14_-_48103207 | 1.28 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr13_-_39160018 | 1.28 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr23_+_44634187 | 1.28 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr7_+_59649399 | 1.27 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr2_+_58377395 | 1.27 |
ENSDART00000193511
|
vapal
|
VAMP (vesicle-associated membrane protein)-associated protein A, like |
| chr4_+_34119824 | 1.26 |
ENSDART00000165637
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr3_-_31618037 | 1.26 |
ENSDART00000183547
|
moto
|
minamoto |
| chr7_-_40578733 | 1.26 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr8_-_50132860 | 1.24 |
ENSDART00000149964
|
antxr1a
|
anthrax toxin receptor 1a |
| chr1_+_57050899 | 1.24 |
ENSDART00000152601
|
si:ch211-1f22.14
|
si:ch211-1f22.14 |
| chr18_-_127558 | 1.24 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr14_-_34700633 | 1.24 |
ENSDART00000150358
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr13_+_13668991 | 1.24 |
ENSDART00000148266
|
pimr48
|
Pim proto-oncogene, serine/threonine kinase, related 48 |
| chr19_+_32158010 | 1.23 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr15_+_19797918 | 1.21 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr16_-_29146624 | 1.21 |
ENSDART00000159814
ENSDART00000009826 |
mef2d
|
myocyte enhancer factor 2d |
| chr1_+_24387659 | 1.21 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr16_-_1503023 | 1.19 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr22_-_10774735 | 1.18 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr20_+_6535438 | 1.17 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr5_+_43870389 | 1.17 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr10_-_29272707 | 1.16 |
ENSDART00000022605
ENSDART00000184594 ENSDART00000185371 |
tmem126a
|
transmembrane protein 126A |
| chr2_-_25143373 | 1.15 |
ENSDART00000160108
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr4_+_64921810 | 1.15 |
ENSDART00000157654
|
BX928743.1
|
|
| chr8_+_32742650 | 1.15 |
ENSDART00000138117
|
hmcn2
|
hemicentin 2 |
| chr12_+_22870372 | 1.15 |
ENSDART00000182264
ENSDART00000191789 ENSDART00000130594 ENSDART00000190433 |
afap1
|
actin filament associated protein 1 |
| chr4_-_56157199 | 1.14 |
ENSDART00000169806
|
si:ch211-207e19.15
|
si:ch211-207e19.15 |
| chr2_+_35880600 | 1.12 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr5_+_9246018 | 1.11 |
ENSDART00000081769
ENSDART00000183189 |
susd1
|
sushi domain containing 1 |
| chr22_-_38274188 | 1.11 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr13_+_713941 | 1.10 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr13_-_39159810 | 1.10 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr20_-_49650293 | 1.09 |
ENSDART00000151239
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr1_+_57176256 | 1.08 |
ENSDART00000152741
|
si:dkey-27j5.6
|
si:dkey-27j5.6 |
| chr22_-_14346575 | 1.08 |
ENSDART00000136326
|
si:dkeyp-122a9.1
|
si:dkeyp-122a9.1 |
| chr2_-_7666021 | 1.07 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr19_-_27564980 | 1.06 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr4_+_38981587 | 1.06 |
ENSDART00000142713
|
si:dkey-66k12.3
|
si:dkey-66k12.3 |
| chr4_-_64605318 | 1.05 |
ENSDART00000170391
|
znf1099
|
zinc finger protein 1099 |
| chr9_+_35077546 | 1.05 |
ENSDART00000142688
|
si:dkey-192g7.3
|
si:dkey-192g7.3 |
| chr21_+_38089036 | 1.03 |
ENSDART00000147219
|
klf8
|
Kruppel-like factor 8 |
| chr8_+_4745340 | 1.02 |
ENSDART00000182894
|
ufd1l
|
ubiquitin recognition factor in ER associated degradation 1 |
| chr19_-_25005609 | 1.01 |
ENSDART00000151129
|
xkr8.2
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 2 |
| chr24_-_21913426 | 1.00 |
ENSDART00000081178
|
c1qtnf9
|
C1q and TNF related 9 |
| chr23_+_19790962 | 1.00 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr7_-_35393900 | 1.00 |
ENSDART00000173769
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr19_-_3741602 | 1.00 |
ENSDART00000170301
|
btr22
|
bloodthirsty-related gene family, member 22 |
| chr22_+_25184459 | 0.99 |
ENSDART00000105308
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr14_-_29859067 | 0.99 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr19_+_7604527 | 0.99 |
ENSDART00000132107
|
s100s
|
S100 calcium binding protein S |
| chr1_+_23783349 | 0.97 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr9_-_33121535 | 0.97 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr23_+_27912079 | 0.97 |
ENSDART00000171859
|
BX539336.1
|
|
| chr12_+_48241841 | 0.97 |
ENSDART00000168616
|
ppa1a
|
pyrophosphatase (inorganic) 1a |
| chr1_+_58094551 | 0.96 |
ENSDART00000146316
|
si:ch211-114l13.1
|
si:ch211-114l13.1 |
| chr3_+_22432069 | 0.96 |
ENSDART00000123054
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr6_-_39270851 | 0.96 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 15.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 1.9 | 5.7 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 1.6 | 9.9 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.4 | 10.8 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 1.0 | 12.9 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.9 | 2.8 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.7 | 2.2 | GO:1905133 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.6 | 14.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.6 | 5.1 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.6 | 10.0 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.4 | 1.3 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.4 | 2.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.4 | 3.4 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.4 | 1.3 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.4 | 2.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.4 | 1.6 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.4 | 4.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 1.2 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.4 | 10.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.4 | 2.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 7.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.3 | 3.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 1.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 1.0 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.3 | 2.3 | GO:1902902 | negative regulation of autophagosome maturation(GO:1901097) negative regulation of autophagosome assembly(GO:1902902) |
| 0.3 | 1.0 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 3.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.3 | 3.0 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 2.3 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.3 | 1.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 0.8 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.3 | 2.8 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 1.3 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.3 | 4.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.2 | 4.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 2.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 1.5 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 1.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 1.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.2 | 1.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 1.0 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.2 | 0.7 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 7.9 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.2 | 1.2 | GO:0043589 | skin morphogenesis(GO:0043589) mesenchymal cell migration(GO:0090497) |
| 0.2 | 0.5 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.2 | 0.6 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 | 0.6 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 12.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.4 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 4.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 3.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 13.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.4 | GO:0033335 | anal fin development(GO:0033335) |
| 0.1 | 1.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 1.2 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 4.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 2.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.8 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 2.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.8 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 2.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.5 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 4.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.1 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 1.0 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.4 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 0.2 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 1.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 12.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 0.8 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 0.4 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.5 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 2.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.7 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 1.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 2.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.7 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 1.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.2 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 1.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 2.4 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 1.1 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 1.4 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.0 | 1.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0097107 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 4.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 2.0 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 1.4 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 7.1 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 1.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.2 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) arachidonate transport(GO:1903963) |
| 0.0 | 1.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.3 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 5.8 | GO:0007005 | mitochondrion organization(GO:0007005) |
| 0.0 | 0.8 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.1 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 1.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.6 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 1.0 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.8 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.6 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 1.8 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.5 | GO:0051085 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.6 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 3.4 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 0.5 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.8 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.5 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.4 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 5.4 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.5 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.7 | 2.8 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.6 | 4.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 2.7 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.5 | 10.5 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 1.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.4 | 0.9 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.4 | 1.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 16.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.4 | 1.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 2.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 9.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 1.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 4.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 7.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.2 | 8.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 2.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 1.8 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.2 | 3.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 2.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 9.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 3.3 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 2.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 2.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 4.5 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.9 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 1.0 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.5 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 1.3 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.0 | 0.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.3 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.5 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 1.8 | 14.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.6 | 7.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 1.4 | 15.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.2 | 3.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.0 | 14.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.0 | 12.9 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.8 | 4.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.5 | 2.6 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.4 | 1.3 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.4 | 1.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 2.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 1.4 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.3 | 9.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 1.7 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 1.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 1.2 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.3 | 1.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.3 | 2.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 2.8 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 4.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 2.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 1.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 2.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 4.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.6 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 1.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 1.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.1 | 1.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 3.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 1.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 24.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 0.6 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 9.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 5.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 1.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 8.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 8.0 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 4.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 3.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.7 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.8 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 9.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 1.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 3.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 5.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 2.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 1.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 3.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 3.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |