Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
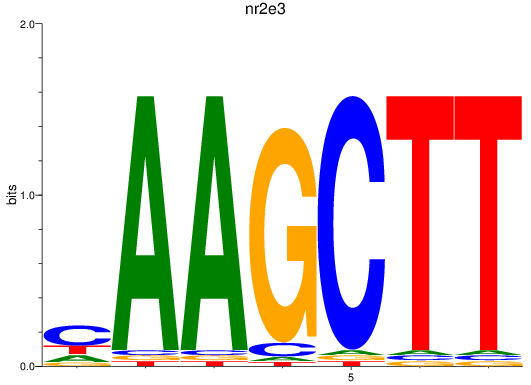
Results for nr2e3
Z-value: 1.10
Transcription factors associated with nr2e3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2e3
|
ENSDARG00000045904 | nuclear receptor subfamily 2, group E, member 3 |
|
nr2e3
|
ENSDARG00000113178 | nuclear receptor subfamily 2, group E, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2e3 | dr11_v1_chr25_+_22274642_22274642 | 0.17 | 9.7e-02 | Click! |
Activity profile of nr2e3 motif
Sorted Z-values of nr2e3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_47633438 | 16.35 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr13_+_27316934 | 14.89 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr3_-_34801041 | 9.80 |
ENSDART00000103043
|
nsfa
|
N-ethylmaleimide-sensitive factor a |
| chr12_-_10275320 | 8.57 |
ENSDART00000170078
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr11_-_1024483 | 8.13 |
ENSDART00000017551
|
slc6a1b
|
solute carrier family 6 (neurotransmitter transporter), member 1b |
| chr3_-_28665291 | 7.93 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr24_-_29997145 | 7.60 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr24_+_39158283 | 7.54 |
ENSDART00000053139
|
atp6v0cb
|
ATPase H+ transporting V0 subunit cb |
| chr14_-_21618005 | 7.45 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr2_+_47582681 | 7.40 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr6_+_38427357 | 7.29 |
ENSDART00000148678
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr10_-_7974155 | 6.99 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr20_+_34913069 | 6.89 |
ENSDART00000007584
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr13_+_27316632 | 6.75 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr20_-_31905968 | 6.71 |
ENSDART00000142806
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr21_+_25643880 | 6.69 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr8_+_16025554 | 6.43 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr3_+_15271943 | 6.42 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr25_-_12203952 | 6.14 |
ENSDART00000158204
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr23_+_19564392 | 6.06 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr6_+_38381957 | 5.93 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr8_+_31941650 | 5.85 |
ENSDART00000138217
|
htr1aa
|
5-hydroxytryptamine (serotonin) receptor 1A a |
| chr10_+_21776911 | 5.77 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr14_-_2202652 | 5.77 |
ENSDART00000171316
|
si:dkey-262j3.7
|
si:dkey-262j3.7 |
| chr24_+_2519761 | 5.73 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr13_+_38520927 | 5.71 |
ENSDART00000137299
ENSDART00000111080 |
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr8_+_3820134 | 5.67 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr24_-_24163201 | 5.62 |
ENSDART00000140170
|
map7d2b
|
MAP7 domain containing 2b |
| chr18_-_38087875 | 5.53 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr23_-_29502287 | 5.41 |
ENSDART00000141075
ENSDART00000053807 |
kif1b
|
kinesin family member 1B |
| chr22_+_34701848 | 5.14 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr12_-_10705916 | 5.03 |
ENSDART00000164038
|
FO704786.1
|
|
| chr21_+_34976600 | 5.01 |
ENSDART00000191672
ENSDART00000139635 |
rbm11
|
RNA binding motif protein 11 |
| chr14_-_2163454 | 5.00 |
ENSDART00000160123
ENSDART00000169653 |
pcdh2ab9
pcdh2ac
|
protocadherin 2 alpha b 9 protocadherin 2 alpha c |
| chr6_+_38427570 | 4.97 |
ENSDART00000170612
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr25_+_7670683 | 4.92 |
ENSDART00000040275
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr2_-_32637592 | 4.85 |
ENSDART00000136353
|
si:dkeyp-73d8.8
|
si:dkeyp-73d8.8 |
| chr20_-_34868814 | 4.61 |
ENSDART00000153049
|
stmn4
|
stathmin-like 4 |
| chr13_+_16522608 | 4.56 |
ENSDART00000182838
ENSDART00000143200 |
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr5_+_36768674 | 4.44 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr3_-_43821381 | 4.43 |
ENSDART00000166021
|
snn
|
stannin |
| chr8_-_23416362 | 4.41 |
ENSDART00000063005
|
gpr173
|
G protein-coupled receptor 173 |
| chr5_-_28606916 | 4.40 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr4_-_5019113 | 4.39 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr2_+_34767171 | 4.38 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr9_-_12034444 | 4.38 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr2_+_47582488 | 4.34 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr1_+_32521469 | 4.29 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr10_+_22782522 | 4.26 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr9_-_3671911 | 4.16 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr3_-_27868183 | 4.16 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr20_+_19238382 | 4.16 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr10_+_24504464 | 4.15 |
ENSDART00000141387
|
mtus2a
|
microtubule associated tumor suppressor candidate 2a |
| chr22_+_20208185 | 4.15 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr9_-_6661657 | 4.13 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr5_+_58397646 | 4.10 |
ENSDART00000180759
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr16_-_9383629 | 4.09 |
ENSDART00000084264
ENSDART00000166958 |
adcy2a
|
adenylate cyclase 2a |
| chr9_+_17348745 | 4.07 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr18_-_38088099 | 4.05 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr6_-_29143463 | 4.01 |
ENSDART00000087440
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr2_+_50608099 | 4.00 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr16_+_5774977 | 4.00 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr10_+_34685135 | 3.94 |
ENSDART00000184999
|
nbeaa
|
neurobeachin a |
| chr12_+_25600685 | 3.93 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr12_+_26632448 | 3.88 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr25_-_7686201 | 3.84 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr4_+_4816695 | 3.82 |
ENSDART00000136962
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr20_-_34453623 | 3.82 |
ENSDART00000005609
|
kifap3a
|
kinesin-associated protein 3a |
| chr6_+_55277419 | 3.82 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr3_+_7617353 | 3.79 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr20_-_44576949 | 3.72 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr25_-_7925019 | 3.70 |
ENSDART00000183309
|
glcea
|
glucuronic acid epimerase a |
| chr8_-_50888806 | 3.69 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr8_+_29856013 | 3.69 |
ENSDART00000061981
ENSDART00000149610 |
hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr6_-_12314475 | 3.69 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr23_+_2778813 | 3.65 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr7_+_13418812 | 3.62 |
ENSDART00000191905
ENSDART00000091567 |
dagla
|
diacylglycerol lipase, alpha |
| chr6_-_13408680 | 3.60 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr14_-_46617228 | 3.59 |
ENSDART00000161010
ENSDART00000171054 |
prom1a
|
prominin 1a |
| chr3_-_13147310 | 3.59 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr5_+_38263240 | 3.58 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr21_+_32332616 | 3.57 |
ENSDART00000154105
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr24_+_15020402 | 3.56 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr16_-_12914288 | 3.55 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr9_+_42063906 | 3.53 |
ENSDART00000048893
|
pcbp3
|
poly(rC) binding protein 3 |
| chr21_-_42202792 | 3.49 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr6_+_39222598 | 3.49 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr19_+_15571290 | 3.48 |
ENSDART00000131134
|
foxo6b
|
forkhead box O6 b |
| chr18_-_7342950 | 3.48 |
ENSDART00000081356
|
caps2
|
calcyphosine 2 |
| chr3_-_43821191 | 3.46 |
ENSDART00000186816
|
snn
|
stannin |
| chr19_+_42231431 | 3.43 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr14_-_43000836 | 3.43 |
ENSDART00000162714
|
pcdh10b
|
protocadherin 10b |
| chr10_+_7182168 | 3.40 |
ENSDART00000172766
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr20_-_50239406 | 3.40 |
ENSDART00000128053
|
tmem121ab
|
transmembrane protein 121Ab |
| chr5_+_32009080 | 3.40 |
ENSDART00000186885
|
scai
|
suppressor of cancer cell invasion |
| chrM_+_8894 | 3.36 |
ENSDART00000093611
|
mt-atp8
|
ATP synthase 8, mitochondrial |
| chr16_-_13004166 | 3.30 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr14_+_24215046 | 3.28 |
ENSDART00000079215
|
stc2a
|
stanniocalcin 2a |
| chr17_-_3986236 | 3.18 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr9_-_31596016 | 3.18 |
ENSDART00000142289
|
nalcn
|
sodium leak channel, non-selective |
| chr1_-_50167792 | 3.16 |
ENSDART00000182993
|
BX537118.1
|
|
| chr9_+_6079528 | 3.14 |
ENSDART00000142167
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr10_+_15255198 | 3.13 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr3_+_50312422 | 3.11 |
ENSDART00000157689
|
gas7a
|
growth arrest-specific 7a |
| chr6_+_49021703 | 3.11 |
ENSDART00000149394
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr14_-_2036604 | 3.08 |
ENSDART00000192446
|
BX005294.2
|
|
| chr13_+_22659153 | 3.07 |
ENSDART00000143906
ENSDART00000140472 ENSDART00000078877 ENSDART00000182469 |
sncga
|
synuclein, gamma a |
| chr10_+_15255012 | 3.07 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr5_-_30588539 | 3.05 |
ENSDART00000086686
ENSDART00000159163 ENSDART00000142958 |
c2cd2l
|
c2cd2-like |
| chr9_-_35334642 | 3.04 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr7_-_35710263 | 3.04 |
ENSDART00000043857
|
irx5a
|
iroquois homeobox 5a |
| chr23_-_17657348 | 3.04 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr2_-_8017579 | 3.00 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr12_+_42436328 | 3.00 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr18_-_29962234 | 2.99 |
ENSDART00000144996
|
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr14_+_32022272 | 2.99 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr6_-_35310224 | 2.95 |
ENSDART00000148997
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr6_-_52400896 | 2.95 |
ENSDART00000187624
|
mmp24
|
matrix metallopeptidase 24 |
| chr9_-_35557397 | 2.95 |
ENSDART00000100681
|
ncam2
|
neural cell adhesion molecule 2 |
| chr22_-_3595439 | 2.94 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr18_-_14734678 | 2.94 |
ENSDART00000142462
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr1_-_9980765 | 2.93 |
ENSDART00000142906
|
si:dkeyp-75b4.7
|
si:dkeyp-75b4.7 |
| chr6_-_46403475 | 2.91 |
ENSDART00000154148
|
camk1a
|
calcium/calmodulin-dependent protein kinase Ia |
| chr9_-_4506819 | 2.89 |
ENSDART00000113975
|
kcnj3a
|
potassium inwardly-rectifying channel, subfamily J, member 3a |
| chr1_-_42778510 | 2.84 |
ENSDART00000190172
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr8_-_14080534 | 2.83 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr8_+_14855471 | 2.81 |
ENSDART00000134775
|
si:dkey-21p1.3
|
si:dkey-21p1.3 |
| chr2_+_45382433 | 2.79 |
ENSDART00000142251
|
wdr47a
|
WD repeat domain 47a |
| chr21_+_10794914 | 2.76 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr15_-_34213898 | 2.75 |
ENSDART00000191945
ENSDART00000186089 |
etv1
|
ets variant 1 |
| chr7_-_34339845 | 2.74 |
ENSDART00000173816
|
madd
|
MAP-kinase activating death domain |
| chr12_+_15622621 | 2.74 |
ENSDART00000079784
|
plcd3b
|
phospholipase C, delta 3b |
| chr12_+_20667301 | 2.73 |
ENSDART00000144804
|
mxra7
|
matrix-remodelling associated 7 |
| chr9_+_10014817 | 2.73 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr2_-_37043540 | 2.72 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr14_-_30724165 | 2.69 |
ENSDART00000020936
|
fibpa
|
fibroblast growth factor (acidic) intracellular binding protein a |
| chr8_-_15398760 | 2.67 |
ENSDART00000162011
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr15_+_7992906 | 2.66 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr5_-_10946232 | 2.66 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr9_-_40935934 | 2.63 |
ENSDART00000155604
|
C2orf88
|
Small membrane A-kinase anchor protein |
| chr18_-_8857137 | 2.60 |
ENSDART00000126331
|
prrt4
|
proline-rich transmembrane protein 4 |
| chr18_+_50276653 | 2.59 |
ENSDART00000192120
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr3_+_24511959 | 2.53 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr19_-_27564980 | 2.49 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr19_+_37925616 | 2.48 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr20_-_16849306 | 2.47 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr15_-_28107502 | 2.47 |
ENSDART00000152270
ENSDART00000167129 |
cryba1a
|
crystallin, beta A1a |
| chr6_-_53885514 | 2.46 |
ENSDART00000173812
ENSDART00000127144 |
cacna2d2a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2a |
| chr8_+_7097740 | 2.43 |
ENSDART00000159670
|
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr24_+_8904741 | 2.43 |
ENSDART00000140924
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr20_-_28698172 | 2.39 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr10_-_27009413 | 2.39 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr4_+_68476490 | 2.38 |
ENSDART00000168543
|
si:dkey-237g15.2
|
si:dkey-237g15.2 |
| chr9_+_17982737 | 2.38 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr9_+_17983463 | 2.37 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr17_+_18117029 | 2.36 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr4_+_5848229 | 2.34 |
ENSDART00000161101
ENSDART00000067357 |
lyrm5a
|
LYR motif containing 5a |
| chr4_+_23223881 | 2.33 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr22_+_4707663 | 2.32 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr8_+_13364950 | 2.32 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr23_+_26491931 | 2.31 |
ENSDART00000190124
ENSDART00000132905 |
si:ch73-206d17.1
|
si:ch73-206d17.1 |
| chr1_-_54997746 | 2.31 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr10_+_45128375 | 2.30 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr11_+_14622379 | 2.28 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr7_-_35708450 | 2.27 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr3_+_59851537 | 2.27 |
ENSDART00000180997
|
CU693479.1
|
|
| chr5_-_20135679 | 2.27 |
ENSDART00000079402
|
usp30
|
ubiquitin specific peptidase 30 |
| chr21_+_10076203 | 2.25 |
ENSDART00000190383
|
CABZ01043506.1
|
|
| chr3_+_25503442 | 2.25 |
ENSDART00000190117
|
mchr1b
|
melanin-concentrating hormone receptor 1b |
| chr4_-_5018705 | 2.23 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr9_+_22632126 | 2.23 |
ENSDART00000139434
|
etv5a
|
ets variant 5a |
| chr11_+_13071645 | 2.22 |
ENSDART00000162259
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr12_+_26471712 | 2.21 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr6_-_25189739 | 2.20 |
ENSDART00000181980
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr18_+_11970987 | 2.20 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr23_+_21459263 | 2.20 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr17_+_18117358 | 2.18 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr1_+_22851745 | 2.18 |
ENSDART00000138235
ENSDART00000016488 |
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr3_+_51563695 | 2.17 |
ENSDART00000008607
|
ttyh2l
|
tweety homolog 2, like |
| chr22_+_31930650 | 2.17 |
ENSDART00000092090
|
dock3
|
dedicator of cytokinesis 3 |
| chr5_-_50992690 | 2.16 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr17_-_2721336 | 2.16 |
ENSDART00000109582
ENSDART00000192691 ENSDART00000189381 |
spata7
|
spermatogenesis associated 7 |
| chr16_-_9869056 | 2.15 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr7_-_5487593 | 2.14 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr14_-_46213571 | 2.13 |
ENSDART00000056734
|
setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_-_22851481 | 2.10 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr23_+_21479958 | 2.10 |
ENSDART00000188302
ENSDART00000144320 |
si:dkey-1c11.1
|
si:dkey-1c11.1 |
| chr4_-_8902406 | 2.10 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr12_+_2631711 | 2.10 |
ENSDART00000114209
|
gdf10b
|
growth differentiation factor 10b |
| chr20_-_15132151 | 2.09 |
ENSDART00000063884
|
si:dkey-239i20.4
|
si:dkey-239i20.4 |
| chr16_+_25068576 | 2.08 |
ENSDART00000125838
|
im:7147486
|
im:7147486 |
| chr23_+_19202636 | 2.05 |
ENSDART00000162797
|
si:dkey-109j17.5
|
si:dkey-109j17.5 |
| chr19_-_5812319 | 2.04 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr9_+_10014514 | 2.03 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr10_+_33902264 | 2.03 |
ENSDART00000193793
|
fryb
|
furry homolog b (Drosophila) |
| chr23_-_35066816 | 2.02 |
ENSDART00000168731
ENSDART00000163731 |
BX294434.1
|
|
| chr20_+_29690901 | 2.02 |
ENSDART00000142669
|
mboat2b
|
membrane bound O-acyltransferase domain containing 2b |
| chr1_-_47114310 | 2.00 |
ENSDART00000144899
ENSDART00000053157 |
setd4
|
SET domain containing 4 |
| chr7_-_46781614 | 1.99 |
ENSDART00000173891
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr7_+_63325819 | 1.99 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2e3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.8 | GO:0048211 | Golgi vesicle docking(GO:0048211) anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 1.8 | 5.4 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 1.5 | 4.4 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 1.1 | 5.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.9 | 3.6 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.8 | 4.2 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.7 | 3.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.7 | 2.8 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.7 | 4.1 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.6 | 7.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.6 | 3.6 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.5 | 5.4 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.5 | 5.3 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.5 | 4.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.5 | 3.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.5 | 1.9 | GO:0006522 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.4 | 1.7 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.4 | 2.5 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 2.8 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.4 | 7.7 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.4 | 1.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.4 | 6.8 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.4 | 1.6 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.4 | 1.2 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.4 | 2.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.4 | 1.9 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.4 | 3.4 | GO:0021794 | thalamus development(GO:0021794) |
| 0.4 | 1.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.4 | 2.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.4 | 1.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.4 | 6.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.4 | 4.6 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.3 | 1.0 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.3 | 3.7 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.3 | 3.9 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.3 | 0.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 4.6 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.3 | 1.2 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.3 | 22.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.3 | 0.9 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.3 | 1.5 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.3 | 0.6 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.3 | 23.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.3 | 0.8 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 3.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 5.8 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.3 | 2.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.3 | 3.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 0.8 | GO:0021985 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.2 | 3.7 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.2 | 4.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 3.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 1.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.2 | 1.4 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.2 | 19.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.2 | 3.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 0.9 | GO:0010259 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.9 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.2 | 4.9 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 3.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 0.9 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.2 | 1.8 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.2 | 2.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 1.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 0.6 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.2 | 0.6 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) pronuclear migration(GO:0035046) |
| 0.2 | 2.9 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 11.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 0.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 2.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 12.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.2 | 2.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 6.7 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.2 | 1.3 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.2 | 0.9 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 0.6 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 0.8 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 1.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 0.6 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 1.0 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 3.0 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 1.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 4.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 6.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 1.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 3.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.9 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 1.3 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 3.4 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
| 0.1 | 1.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 3.4 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.5 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 2.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 3.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 0.9 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 4.4 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.1 | 2.3 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 4.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 1.9 | GO:0050890 | cognition(GO:0050890) |
| 0.1 | 0.7 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.1 | 1.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.3 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 5.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 0.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 4.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 4.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.8 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.3 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 1.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.9 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.7 | GO:0060324 | face development(GO:0060324) |
| 0.1 | 5.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 17.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 4.3 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.1 | 7.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 2.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.8 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 13.0 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 0.7 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 2.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 2.4 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 2.1 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.7 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 2.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase II promoter(GO:0042795) snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 1.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 1.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 3.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 1.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.2 | GO:0035775 | pronephric glomerulus morphogenesis(GO:0035775) glomerulus morphogenesis(GO:0072102) |
| 0.1 | 1.0 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.1 | 0.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.4 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 0.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.3 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 1.0 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 0.4 | GO:0060114 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 1.1 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.1 | 0.8 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 2.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 2.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.0 | 0.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 1.7 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 2.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.6 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 4.3 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 2.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 3.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 10.6 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.9 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 3.0 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.3 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 3.0 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 4.4 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.7 | GO:0046834 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 3.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.8 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.2 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.2 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 2.8 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.7 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 1.7 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 5.2 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.5 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 1.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 4.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.0 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.2 | GO:0002521 | leukocyte differentiation(GO:0002521) |
| 0.0 | 4.1 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 5.0 | GO:0007420 | brain development(GO:0007420) |
| 0.0 | 0.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) organic acid transmembrane transport(GO:1903825) carboxylic acid transmembrane transport(GO:1905039) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.1 | 6.9 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.0 | 6.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 1.0 | 5.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.9 | 12.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.6 | 21.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.6 | 7.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.6 | 2.8 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.5 | 7.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.4 | 3.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.4 | 6.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.4 | 3.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 3.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 3.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.3 | 1.4 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.2 | 4.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 0.9 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.2 | 0.8 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 2.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 0.6 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.2 | 1.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 7.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 0.8 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.2 | 0.6 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.2 | 2.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 2.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 6.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 13.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 6.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 7.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 9.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 11.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 3.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 11.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 1.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 5.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.4 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.3 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.1 | 2.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 5.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 4.3 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.5 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 3.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 19.2 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 1.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 5.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 11.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 7.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 7.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.1 | 13.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 1.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 0.8 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.0 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 2.9 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 8.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 21.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 2.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 3.4 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 0.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.5 | 6.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 1.0 | 3.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.0 | 6.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.9 | 3.7 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.8 | 2.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.8 | 4.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.8 | 2.3 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.6 | 21.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.6 | 8.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.5 | 1.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.5 | 3.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 21.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.5 | 1.9 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.5 | 1.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.5 | 1.4 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.4 | 2.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.4 | 1.6 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.4 | 6.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.4 | 3.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.4 | 1.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 3.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.3 | 3.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 2.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.3 | 0.9 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 7.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.3 | 3.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.3 | 11.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 7.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 4.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 1.0 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.3 | 2.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 3.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 1.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 0.8 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.2 | 1.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 2.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 3.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.2 | 1.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 5.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 1.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 1.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 1.8 | GO:0016933 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 0.8 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.2 | 0.6 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.2 | 2.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 0.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 3.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 3.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 2.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 5.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.2 | 5.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 0.6 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.2 | 1.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 1.9 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 2.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.8 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 4.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 3.2 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 5.1 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 11.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 2.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 6.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.4 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 1.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 3.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.5 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.9 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 2.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.5 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 2.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 9.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 4.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 3.4 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 2.5 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 1.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 3.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 2.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.1 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 4.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.5 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 2.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.0 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 1.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 2.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.3 | GO:0030060 | malate dehydrogenase activity(GO:0016615) L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 3.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.1 | GO:0009931 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.6 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 2.2 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 5.3 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 2.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.3 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 11.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 2.6 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 1.0 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 5.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.7 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.9 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 4.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.1 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 32.1 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.4 | 7.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 4.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 3.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 4.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 2.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 4.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.9 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 6.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 18.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.9 | 3.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.6 | 9.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.5 | 4.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 3.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 4.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 5.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 1.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 2.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 1.0 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 1.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 1.7 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 4.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.8 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 1.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.5 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.9 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.8 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |