Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
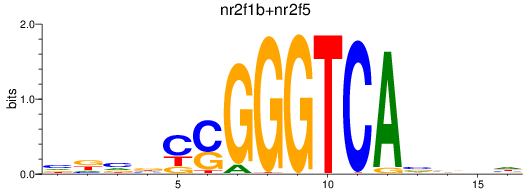
Results for nr2f1b+nr2f5
Z-value: 0.75
Transcription factors associated with nr2f1b+nr2f5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f1b
|
ENSDARG00000017168 | nuclear receptor subfamily 2, group F, member 1b |
|
nr2f5
|
ENSDARG00000033172 | nuclear receptor subfamily 2, group F, member 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f1b | dr11_v1_chr10_+_43797130_43797130 | -0.74 | 7.4e-18 | Click! |
| nr2f5 | dr11_v1_chr16_+_46294337_46294337 | 0.08 | 4.3e-01 | Click! |
Activity profile of nr2f1b+nr2f5 motif
Sorted Z-values of nr2f1b+nr2f5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_56229674 | 10.21 |
ENSDART00000127714
|
c3a.2
|
complement component c3a, duplicate 2 |
| chr20_+_25581627 | 9.74 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
| chr21_+_28445052 | 7.78 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr18_+_8340886 | 7.25 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr19_+_8606883 | 6.32 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr24_+_42149453 | 6.22 |
ENSDART00000128766
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr25_+_1549838 | 6.16 |
ENSDART00000115001
|
avpr1aa
|
arginine vasopressin receptor 1Aa |
| chr4_+_1530287 | 5.81 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr18_-_49283058 | 5.69 |
ENSDART00000076554
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr9_-_9980704 | 5.51 |
ENSDART00000130243
ENSDART00000193475 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr11_+_6010177 | 5.23 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr3_-_19517462 | 5.10 |
ENSDART00000162027
|
CU571315.2
|
|
| chr13_+_22249636 | 5.04 |
ENSDART00000108472
ENSDART00000173123 |
synpo2la
|
synaptopodin 2-like a |
| chr5_+_64856666 | 5.03 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr22_+_37888249 | 4.75 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr22_-_26834043 | 4.67 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr13_-_13754091 | 4.52 |
ENSDART00000131255
|
ky
|
kyphoscoliosis peptidase |
| chr24_+_42149296 | 4.49 |
ENSDART00000154231
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr17_+_5351922 | 4.41 |
ENSDART00000105394
|
runx2a
|
runt-related transcription factor 2a |
| chr18_-_11729 | 4.28 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr20_+_53577502 | 4.24 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr14_+_2487672 | 4.23 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr24_-_22015751 | 3.98 |
ENSDART00000147055
|
si:rp71-1f1.6
|
si:rp71-1f1.6 |
| chr4_-_77120928 | 3.95 |
ENSDART00000174154
|
CU467646.1
|
|
| chr10_-_76352 | 3.84 |
ENSDART00000186560
ENSDART00000144722 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr24_-_22003031 | 3.79 |
ENSDART00000133627
|
si:rp71-1f1.5
|
si:rp71-1f1.5 |
| chr15_+_22394074 | 3.73 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr24_+_42132962 | 3.53 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr14_-_5817039 | 3.45 |
ENSDART00000131820
|
kazald2
|
Kazal-type serine peptidase inhibitor domain 2 |
| chr7_+_27251376 | 3.37 |
ENSDART00000173521
ENSDART00000173962 |
sox6
|
SRY (sex determining region Y)-box 6 |
| chr7_+_4474880 | 3.34 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr19_+_26828502 | 3.30 |
ENSDART00000183654
ENSDART00000126006 ENSDART00000185341 |
FP085398.1
|
|
| chr24_+_42127983 | 3.20 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr2_+_52847049 | 3.06 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr1_+_144284 | 2.96 |
ENSDART00000064061
|
prozb
|
protein Z, vitamin K-dependent plasma glycoprotein b |
| chr24_+_42117231 | 2.79 |
ENSDART00000067734
|
snx16
|
sorting nexin 16 |
| chr15_+_34933552 | 2.76 |
ENSDART00000155368
|
zgc:66024
|
zgc:66024 |
| chr19_+_7938121 | 2.66 |
ENSDART00000140053
ENSDART00000146649 |
si:dkey-266f7.5
|
si:dkey-266f7.5 |
| chr20_-_16906623 | 2.47 |
ENSDART00000012859
ENSDART00000171628 |
psma6b
|
proteasome subunit alpha 6b |
| chr8_-_13419049 | 2.35 |
ENSDART00000133656
|
pimr101
|
Pim proto-oncogene, serine/threonine kinase, related 101 |
| chr4_-_837768 | 2.34 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr21_+_5882300 | 2.31 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr15_-_41756454 | 2.28 |
ENSDART00000139498
ENSDART00000099302 |
ftr72
|
finTRIM family, member 72 |
| chr8_-_13428740 | 2.26 |
ENSDART00000131826
|
CR354547.3
|
|
| chr4_-_13613148 | 2.23 |
ENSDART00000067164
ENSDART00000111247 |
irf5
|
interferon regulatory factor 5 |
| chr8_-_13471916 | 2.23 |
ENSDART00000146558
|
pimr105
|
Pim proto-oncogene, serine/threonine kinase, related 105 |
| chr22_+_344763 | 2.17 |
ENSDART00000181934
|
CU914780.1
|
|
| chr1_-_55785722 | 2.17 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
| chr17_-_53282539 | 2.12 |
ENSDART00000164953
|
FO704905.1
|
|
| chr13_-_50546634 | 2.05 |
ENSDART00000192127
|
CU570781.2
|
|
| chr8_-_13454281 | 2.02 |
ENSDART00000141959
|
CR354547.2
|
|
| chr20_+_43925266 | 1.96 |
ENSDART00000037379
|
clic5b
|
chloride intracellular channel 5b |
| chr1_-_20824411 | 1.96 |
ENSDART00000182810
|
tll1
|
tolloid-like 1 |
| chr22_+_18316144 | 1.94 |
ENSDART00000137985
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr1_-_18772241 | 1.89 |
ENSDART00000147645
ENSDART00000133194 ENSDART00000147525 ENSDART00000191493 |
tbc1d1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr24_-_21991770 | 1.87 |
ENSDART00000147296
|
si:rp71-1f1.4
|
si:rp71-1f1.4 |
| chr8_+_23711842 | 1.84 |
ENSDART00000128783
|
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr18_+_7073130 | 1.83 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr5_-_25084318 | 1.83 |
ENSDART00000089339
|
dph7
|
diphthamide biosynthesis 7 |
| chr5_+_54400971 | 1.82 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr4_+_73215536 | 1.79 |
ENSDART00000174290
|
LO018260.2
|
Danio rerio protein NLRC3-like (LOC101883187), mRNA. |
| chr14_+_34558480 | 1.74 |
ENSDART00000075170
|
pttg1
|
pituitary tumor-transforming 1 |
| chr8_-_13486258 | 1.68 |
ENSDART00000137459
|
pimr104
|
Pim proto-oncogene, serine/threonine kinase, related 104 |
| chr8_-_13442871 | 1.63 |
ENSDART00000144887
|
pimr106
|
Pim proto-oncogene, serine/threonine kinase, related 106 |
| chr2_-_5502256 | 1.56 |
ENSDART00000193037
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
| chr15_+_27364394 | 1.44 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr18_-_6975175 | 1.42 |
ENSDART00000134194
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr5_-_8910703 | 1.36 |
ENSDART00000097221
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr17_+_389218 | 1.32 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr13_+_50778187 | 1.28 |
ENSDART00000159597
|
dntt
|
deoxynucleotidyltransferase, terminal |
| chr16_+_20895904 | 1.15 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr6_+_103361 | 1.11 |
ENSDART00000151899
|
ldlrb
|
low density lipoprotein receptor b |
| chr5_-_40292299 | 1.07 |
ENSDART00000097525
|
snx18a
|
sorting nexin 18a |
| chr2_+_33382648 | 0.92 |
ENSDART00000137207
ENSDART00000098831 |
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr15_+_1199407 | 0.89 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr15_+_47582207 | 0.82 |
ENSDART00000159388
|
CABZ01087566.1
|
|
| chr2_+_2904080 | 0.76 |
ENSDART00000110416
|
eif2b5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon |
| chr14_+_46118834 | 0.67 |
ENSDART00000124417
ENSDART00000017785 |
naa15a
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit a |
| chr4_+_9011448 | 0.66 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr2_-_16449504 | 0.61 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr15_-_34933560 | 0.58 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr1_-_26544268 | 0.55 |
ENSDART00000152117
|
si:ch211-202n12.2
|
si:ch211-202n12.2 |
| chr23_+_25822742 | 0.52 |
ENSDART00000184436
|
r3hdml
|
R3H domain containing-like |
| chr7_-_2039060 | 0.48 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr16_+_53710416 | 0.42 |
ENSDART00000155929
|
nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr9_+_500052 | 0.35 |
ENSDART00000166707
|
CU984600.1
|
|
| chr7_-_26446726 | 0.34 |
ENSDART00000173607
|
zgc:172079
|
zgc:172079 |
| chr24_+_37700259 | 0.33 |
ENSDART00000066556
|
zmp:0000000624
|
zmp:0000000624 |
| chr4_+_9011825 | 0.28 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr2_-_1468258 | 0.23 |
ENSDART00000114431
|
gpaa1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr19_-_48039400 | 0.20 |
ENSDART00000166748
ENSDART00000165921 |
csf3b
|
colony stimulating factor 3 (granulocyte) b |
| chr14_+_41518257 | 0.20 |
ENSDART00000050037
|
chrnb3b
|
cholinergic receptor, nicotinic, beta 3b (neuronal) |
| chr3_-_12970418 | 0.10 |
ENSDART00000158747
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr18_+_910992 | 0.09 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr8_-_41273768 | 0.04 |
ENSDART00000108518
|
rnf10
|
ring finger protein 10 |
| chr22_-_31788170 | 0.01 |
ENSDART00000170925
|
pimr207
|
Pim proto-oncogene, serine/threonine kinase, related 207 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f1b+nr2f5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.7 | 4.2 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.6 | 6.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.5 | 2.0 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.5 | 1.4 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.4 | 7.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.3 | 3.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.3 | 0.9 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 2.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 0.9 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 2.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 5.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 1.8 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.2 | 10.2 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.2 | 9.7 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.2 | 5.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 15.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 1.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.6 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 5.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 2.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 4.4 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 1.7 | GO:0033048 | negative regulation of sister chromatid segregation(GO:0033046) negative regulation of mitotic sister chromatid segregation(GO:0033048) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.1 | 3.1 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 6.7 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 3.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.4 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 4.2 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 3.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.9 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 2.2 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 8.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.3 | GO:0006304 | DNA modification(GO:0006304) |
| 0.0 | 3.4 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 2.0 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.3 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 1.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 3.1 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 3.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 2.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 2.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 0.8 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 4.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.9 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 7.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 2.2 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 5.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 4.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 33.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.6 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.6 | 7.8 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 1.0 | 7.3 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.6 | 6.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.5 | 4.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 6.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 3.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 9.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 25.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 3.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 5.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.4 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.1 | 0.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 5.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 4.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 3.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 5.4 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.5 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 5.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 6.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 7.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 5.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 4.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 6.7 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 2.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |