Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
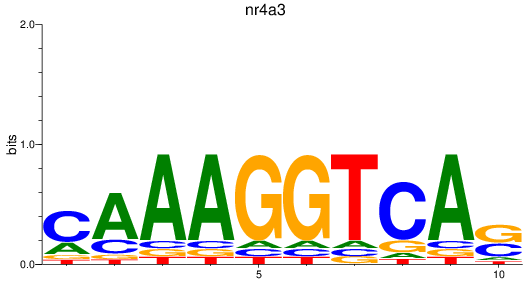
Results for nr4a3
Z-value: 0.81
Transcription factors associated with nr4a3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a3
|
ENSDARG00000055854 | nuclear receptor subfamily 4, group A, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a3 | dr11_v1_chr16_+_27349585_27349585 | 0.17 | 1.0e-01 | Click! |
Activity profile of nr4a3 motif
Sorted Z-values of nr4a3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_1789357 | 10.42 |
ENSDART00000006449
|
atp1a1a.2
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 2 |
| chr7_+_22680560 | 9.14 |
ENSDART00000133761
|
ponzr4
|
plac8 onzin related protein 4 |
| chr5_+_52167986 | 8.28 |
ENSDART00000162256
ENSDART00000073626 |
slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr16_-_31976269 | 7.49 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr22_+_19220459 | 7.18 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr9_+_35077546 | 7.12 |
ENSDART00000142688
|
si:dkey-192g7.3
|
si:dkey-192g7.3 |
| chr25_+_29160102 | 6.50 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr12_+_46634736 | 6.10 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr18_-_48492951 | 5.98 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr1_+_58182400 | 5.92 |
ENSDART00000144784
|
si:ch211-15j1.1
|
si:ch211-15j1.1 |
| chr5_+_29831235 | 5.76 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr22_+_6254194 | 5.71 |
ENSDART00000112388
ENSDART00000135176 |
rnasel4
|
ribonuclease like 4 |
| chr9_-_22821901 | 5.70 |
ENSDART00000101711
|
neb
|
nebulin |
| chr7_+_34297271 | 5.47 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr6_+_18142623 | 5.25 |
ENSDART00000169431
ENSDART00000158841 |
si:dkey-237i9.8
|
si:dkey-237i9.8 |
| chr22_-_7050 | 5.24 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr8_+_1170015 | 5.17 |
ENSDART00000081457
ENSDART00000164116 |
ccl27a
|
chemokine (C-C motif) ligand 27a |
| chr16_-_19568795 | 5.09 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr3_+_31662126 | 5.06 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr3_+_55100045 | 4.95 |
ENSDART00000113098
|
hbaa1
|
hemoglobin, alpha adult 1 |
| chr22_-_28668442 | 4.81 |
ENSDART00000182377
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr10_+_10351685 | 4.80 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr9_+_23770666 | 4.79 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr9_-_22822084 | 4.72 |
ENSDART00000142020
|
neb
|
nebulin |
| chr19_+_11214007 | 4.66 |
ENSDART00000127362
|
si:ch73-109i22.2
|
si:ch73-109i22.2 |
| chr6_+_30533504 | 4.59 |
ENSDART00000155842
|
wwc3
|
WWC family member 3 |
| chr3_+_39566999 | 4.57 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr18_-_16801033 | 4.55 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr22_+_19407531 | 4.54 |
ENSDART00000141060
|
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr20_+_36806398 | 4.46 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr18_-_48550426 | 4.39 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr1_-_52431220 | 4.34 |
ENSDART00000111256
|
zgc:194101
|
zgc:194101 |
| chr19_-_7110617 | 4.25 |
ENSDART00000104838
|
psmb8a
|
proteasome subunit beta 8A |
| chr14_-_17121676 | 4.22 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr4_-_17741513 | 4.21 |
ENSDART00000141680
|
mybpc1
|
myosin binding protein C, slow type |
| chr21_-_22724980 | 3.99 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr20_+_6535438 | 3.92 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr20_-_25369767 | 3.92 |
ENSDART00000180278
|
itsn2a
|
intersectin 2a |
| chr23_+_42434348 | 3.92 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr7_+_13988075 | 3.91 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr10_+_8875195 | 3.91 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr21_+_6034783 | 3.91 |
ENSDART00000169613
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr24_-_26820698 | 3.88 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr21_-_5879897 | 3.88 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr9_+_23772516 | 3.84 |
ENSDART00000183126
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr10_+_24692076 | 3.76 |
ENSDART00000181600
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr12_+_47439 | 3.72 |
ENSDART00000114732
|
mrpl58
|
mitochondrial ribosomal protein L58 |
| chr3_+_30257582 | 3.65 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr20_+_6535176 | 3.63 |
ENSDART00000054652
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr1_+_38153944 | 3.61 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr5_-_30984010 | 3.58 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr16_+_50969248 | 3.49 |
ENSDART00000172068
|
si:dkeyp-97a10.2
|
si:dkeyp-97a10.2 |
| chr5_-_14509137 | 3.48 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr12_+_46791907 | 3.46 |
ENSDART00000110304
|
vcla
|
vinculin a |
| chr7_+_33172066 | 3.43 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr23_+_44614056 | 3.43 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr19_-_1947403 | 3.37 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr24_+_81527 | 3.37 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr20_-_26937453 | 3.29 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr3_-_26921475 | 3.29 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr25_+_7591293 | 3.29 |
ENSDART00000130416
|
FO704779.1
|
|
| chr5_-_38197080 | 3.28 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr23_+_46183410 | 3.27 |
ENSDART00000167596
ENSDART00000151149 ENSDART00000150896 |
btr31
|
bloodthirsty-related gene family, member 31 |
| chr3_-_32590164 | 3.25 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr6_-_35439406 | 3.19 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr3_-_34561624 | 3.18 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr22_-_31020690 | 3.07 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr1_+_24469313 | 3.07 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr8_-_40183197 | 3.04 |
ENSDART00000005118
|
gpx8
|
glutathione peroxidase 8 (putative) |
| chr19_-_35439237 | 3.02 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr7_+_34296789 | 3.02 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr16_-_11859309 | 3.02 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr14_+_21905492 | 2.99 |
ENSDART00000054437
|
tbc1d10c
|
TBC1 domain family, member 10C |
| chr20_-_36887516 | 2.93 |
ENSDART00000076313
|
txlnba
|
taxilin beta a |
| chr2_+_7106837 | 2.92 |
ENSDART00000138691
|
vcam1a
|
vascular cell adhesion molecule 1a |
| chr12_+_14084291 | 2.91 |
ENSDART00000189734
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr13_-_214122 | 2.83 |
ENSDART00000169273
|
ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr21_+_261490 | 2.81 |
ENSDART00000177919
|
jak2a
|
Janus kinase 2a |
| chr12_-_44072716 | 2.77 |
ENSDART00000162509
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr1_+_58353661 | 2.71 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr20_+_37794633 | 2.70 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr2_-_58075414 | 2.68 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr22_+_9806867 | 2.60 |
ENSDART00000137182
ENSDART00000106103 |
si:ch211-236g6.1
|
si:ch211-236g6.1 |
| chr22_-_26005894 | 2.59 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr6_-_35046735 | 2.59 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr8_-_22542467 | 2.47 |
ENSDART00000182588
ENSDART00000134542 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr22_+_987788 | 2.45 |
ENSDART00000149486
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr7_+_14005111 | 2.42 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr9_+_8761099 | 2.39 |
ENSDART00000159077
|
col4a2
|
collagen, type IV, alpha 2 |
| chr22_-_26100282 | 2.36 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr1_-_39859626 | 2.36 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr1_+_58260886 | 2.33 |
ENSDART00000110453
|
si:dkey-222h21.8
|
si:dkey-222h21.8 |
| chr19_+_2631565 | 2.32 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr9_-_42861080 | 2.29 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr16_-_24668620 | 2.29 |
ENSDART00000012807
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr1_+_58067815 | 2.29 |
ENSDART00000156678
|
si:ch211-114l13.4
|
si:ch211-114l13.4 |
| chr1_-_58868306 | 2.27 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr1_-_58975098 | 2.22 |
ENSDART00000189899
|
CABZ01114581.1
|
|
| chr22_-_6923347 | 2.22 |
ENSDART00000082048
|
CABZ01065328.1
|
|
| chr6_-_54111928 | 2.19 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr21_-_25250594 | 2.15 |
ENSDART00000163862
|
nfrkb
|
nuclear factor related to kappaB binding protein |
| chr24_-_28419444 | 2.15 |
ENSDART00000105749
|
nrros
|
negative regulator of reactive oxygen species |
| chr15_+_28268135 | 2.10 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr18_-_46295132 | 2.09 |
ENSDART00000190940
|
pld3
|
phospholipase D family, member 3 |
| chr7_+_24520518 | 2.08 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr7_-_71331690 | 2.08 |
ENSDART00000149682
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr3_-_49925313 | 2.03 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr9_-_9419704 | 2.03 |
ENSDART00000138996
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr14_-_48103207 | 1.98 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr1_+_57050899 | 1.90 |
ENSDART00000152601
|
si:ch211-1f22.14
|
si:ch211-1f22.14 |
| chr24_-_2843107 | 1.88 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr1_-_9644630 | 1.85 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr5_+_36896933 | 1.82 |
ENSDART00000151984
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr20_-_26936887 | 1.82 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr5_+_21891305 | 1.80 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr1_-_43915423 | 1.80 |
ENSDART00000181915
ENSDART00000113673 |
scpp5
|
secretory calcium-binding phosphoprotein 5 |
| chr4_-_77135076 | 1.77 |
ENSDART00000174184
|
zgc:173770
|
zgc:173770 |
| chr10_-_32524771 | 1.76 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr4_-_77116266 | 1.76 |
ENSDART00000174249
|
CU467646.2
|
|
| chr20_+_49119633 | 1.72 |
ENSDART00000151435
|
cd109
|
CD109 molecule |
| chr12_-_4185554 | 1.66 |
ENSDART00000152313
|
si:dkey-32n7.7
|
si:dkey-32n7.7 |
| chr16_+_52343905 | 1.64 |
ENSDART00000131051
|
ifnlr1
|
interferon lambda receptor 1 |
| chr10_-_26202766 | 1.60 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr20_+_49081967 | 1.58 |
ENSDART00000112689
|
crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr3_+_57825938 | 1.57 |
ENSDART00000128815
|
cenpx
|
centromere protein X |
| chr21_+_18405585 | 1.52 |
ENSDART00000139318
|
si:dkey-1d7.3
|
si:dkey-1d7.3 |
| chr10_+_45031398 | 1.52 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr4_+_77957611 | 1.51 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr12_+_45238292 | 1.51 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr8_-_21091961 | 1.50 |
ENSDART00000100281
|
aldh9a1a.2
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 2 |
| chr23_+_5736226 | 1.48 |
ENSDART00000134527
ENSDART00000112220 |
ftr57
|
finTRIM family, member 57 |
| chr3_+_3454610 | 1.47 |
ENSDART00000024900
|
zgc:165453
|
zgc:165453 |
| chr23_+_24124684 | 1.46 |
ENSDART00000144478
|
si:dkey-21o19.2
|
si:dkey-21o19.2 |
| chr25_-_35095129 | 1.46 |
ENSDART00000099866
ENSDART00000099868 |
kif15
|
kinesin family member 15 |
| chr19_-_79202 | 1.45 |
ENSDART00000166009
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr19_+_1465004 | 1.45 |
ENSDART00000159157
|
CABZ01073736.1
|
|
| chr4_-_77130289 | 1.45 |
ENSDART00000174380
|
CU467646.7
|
|
| chr4_-_77120928 | 1.41 |
ENSDART00000174154
|
CU467646.1
|
|
| chr17_+_15213496 | 1.39 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr15_+_11840311 | 1.39 |
ENSDART00000167671
|
prkd2
|
protein kinase D2 |
| chr2_-_17115256 | 1.39 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr17_-_8592824 | 1.39 |
ENSDART00000127022
|
CU462878.1
|
|
| chr4_+_76752707 | 1.38 |
ENSDART00000082121
|
ms4a17a.4
|
membrane-spanning 4-domains, subfamily A, member 17A.4 |
| chr19_-_47997424 | 1.38 |
ENSDART00000081675
|
ctnnb2
|
catenin, beta 2 |
| chr15_-_16946124 | 1.37 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr25_+_7571920 | 1.37 |
ENSDART00000042928
|
fuk
|
fucokinase |
| chr18_+_44769027 | 1.36 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr23_+_42336084 | 1.35 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr6_-_40429411 | 1.35 |
ENSDART00000156005
ENSDART00000156357 |
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr21_-_1640547 | 1.31 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr4_-_77135340 | 1.30 |
ENSDART00000180581
ENSDART00000179901 |
CU467646.7
|
|
| chr7_-_71331499 | 1.30 |
ENSDART00000081245
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr22_-_24818066 | 1.29 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr14_+_31788733 | 1.28 |
ENSDART00000109063
|
cd40lg
|
CD40 ligand |
| chr17_-_45164291 | 1.25 |
ENSDART00000062109
|
noxred1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr22_-_6361178 | 1.25 |
ENSDART00000150147
|
zgc:113298
|
zgc:113298 |
| chr2_+_5793908 | 1.23 |
ENSDART00000145219
|
slc35d1b
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1b |
| chr25_+_9003230 | 1.21 |
ENSDART00000180330
ENSDART00000142917 |
rag1
|
recombination activating gene 1 |
| chr3_+_39568290 | 1.19 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr20_-_38446891 | 1.17 |
ENSDART00000192013
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr6_-_1587291 | 1.16 |
ENSDART00000067592
ENSDART00000178877 |
zgc:123305
|
zgc:123305 |
| chr24_+_13869092 | 1.15 |
ENSDART00000176492
|
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr3_-_3448095 | 1.14 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr8_-_7440364 | 1.13 |
ENSDART00000180659
|
hdac6
|
histone deacetylase 6 |
| chr15_+_5973909 | 1.13 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr3_+_7771420 | 1.13 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr3_+_23047241 | 1.12 |
ENSDART00000103858
|
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr11_+_37278457 | 1.12 |
ENSDART00000188946
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr1_-_59139599 | 1.11 |
ENSDART00000152233
|
si:ch1073-110a20.3
|
si:ch1073-110a20.3 |
| chr24_+_28528000 | 1.09 |
ENSDART00000155924
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr18_-_43866526 | 1.07 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr7_-_5039562 | 1.07 |
ENSDART00000154438
|
ltb4r
|
leukotriene B4 receptor |
| chr11_-_44999858 | 1.07 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr14_+_20941534 | 1.06 |
ENSDART00000185616
|
zgc:66433
|
zgc:66433 |
| chr25_-_31898552 | 1.06 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr10_+_8629275 | 1.06 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr17_+_1514711 | 1.06 |
ENSDART00000165641
|
akt1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr7_-_26924903 | 1.04 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr9_+_500052 | 1.04 |
ENSDART00000166707
|
CU984600.1
|
|
| chr6_+_42818963 | 1.02 |
ENSDART00000184833
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr6_+_30703828 | 1.01 |
ENSDART00000104628
|
insl5a
|
insulin-like 5a |
| chr8_-_27849770 | 1.01 |
ENSDART00000190196
ENSDART00000181244 |
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr7_+_32021982 | 1.00 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr9_+_41914378 | 0.99 |
ENSDART00000130434
ENSDART00000007058 |
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr13_+_7241170 | 0.96 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr22_-_38274188 | 0.95 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr5_+_61556172 | 0.95 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr18_+_38321039 | 0.94 |
ENSDART00000132534
ENSDART00000111260 ENSDART00000192806 |
alx4b
|
ALX homeobox 4b |
| chr11_-_40681011 | 0.94 |
ENSDART00000166372
ENSDART00000110622 ENSDART00000159713 |
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr9_-_44905867 | 0.93 |
ENSDART00000138316
ENSDART00000131252 ENSDART00000179383 ENSDART00000159337 |
zgc:66484
|
zgc:66484 |
| chr16_-_30927454 | 0.93 |
ENSDART00000184015
|
slc45a4
|
solute carrier family 45, member 4 |
| chr19_-_703898 | 0.91 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr7_-_51775688 | 0.90 |
ENSDART00000149793
|
bmp15
|
bone morphogenetic protein 15 |
| chr3_-_34034498 | 0.90 |
ENSDART00000151145
|
ighv11-2
|
immunoglobulin heavy variable 11-2 |
| chr10_-_35108683 | 0.89 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr4_+_15942075 | 0.87 |
ENSDART00000147964
|
si:dkey-117n7.2
|
si:dkey-117n7.2 |
| chr6_-_49873020 | 0.86 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr23_-_16737161 | 0.86 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr15_-_6615555 | 0.84 |
ENSDART00000152725
|
atm
|
ATM serine/threonine kinase |
| chr10_+_26646104 | 0.84 |
ENSDART00000187685
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 1.7 | 10.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.7 | 5.1 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 1.2 | 3.7 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 1.2 | 8.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.1 | 3.4 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 1.0 | 5.0 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.8 | 2.4 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.8 | 3.0 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.7 | 2.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.6 | 3.5 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.5 | 1.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.5 | 6.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.4 | 1.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.4 | 3.4 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.4 | 5.8 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.4 | 10.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.4 | 1.1 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.3 | 1.4 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.3 | 3.3 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.3 | 1.3 | GO:0002639 | regulation of immunoglobulin production(GO:0002637) positive regulation of immunoglobulin production(GO:0002639) |
| 0.3 | 4.0 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.3 | 3.5 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.3 | 1.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 1.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.3 | 0.8 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.3 | 5.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 0.7 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 4.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.4 | GO:0089700 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.2 | 0.9 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.2 | 3.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 3.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 0.9 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 3.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 2.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.7 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 2.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 3.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.8 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 4.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 7.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 1.1 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 1.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 6.5 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 1.4 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.1 | 2.8 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 4.8 | GO:0043491 | protein kinase B signaling(GO:0043491) |
| 0.1 | 1.1 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 1.0 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 2.3 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.7 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 3.7 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 4.5 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.9 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.9 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.5 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 4.2 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 2.0 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.4 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 3.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.7 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.0 | 2.1 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 2.2 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.3 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 1.9 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 2.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.9 | GO:0002685 | regulation of leukocyte migration(GO:0002685) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 1.0 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 2.5 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.8 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.7 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 1.5 | GO:0030203 | glycosaminoglycan metabolic process(GO:0030203) |
| 0.0 | 1.8 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 3.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 7.1 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 1.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 3.9 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 1.1 | GO:0006476 | protein deacetylation(GO:0006476) |
| 0.0 | 1.6 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 3.3 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 1.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.6 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 4.9 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:0003314 | heart rudiment morphogenesis(GO:0003314) |
| 0.0 | 0.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 6.9 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.5 | 2.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.5 | 1.4 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.4 | 3.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 3.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 4.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 1.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 1.2 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.2 | 0.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 1.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 4.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 4.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.5 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 1.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 4.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 10.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 7.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 3.2 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 3.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 13.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 5.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 3.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 2.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 3.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 5.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 12.9 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.2 | 4.8 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 1.1 | 6.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.0 | 10.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.0 | 2.9 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.8 | 3.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.7 | 10.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.7 | 5.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.7 | 7.9 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.6 | 3.8 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.6 | 2.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.6 | 5.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.5 | 2.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.5 | 1.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 2.6 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.5 | 1.4 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.4 | 3.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 4.9 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 2.0 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.4 | 5.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 1.5 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.4 | 3.7 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.4 | 1.1 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.3 | 1.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.3 | 1.2 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.3 | 1.2 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.3 | 1.1 | GO:0047611 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.3 | 3.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 1.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.3 | 2.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.3 | 3.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 4.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 3.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 0.9 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.2 | 1.3 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.2 | 1.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.2 | 0.9 | GO:0050780 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.2 | 1.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 1.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 3.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.7 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.1 | 3.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 5.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 14.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 6.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 3.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 7.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 3.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 3.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 2.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 7.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 3.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 10.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 5.1 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 2.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 5.0 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 2.1 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 4.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.4 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 6.8 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 1.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 3.2 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 3.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 9.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 2.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 8.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.4 | 4.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 5.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 1.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 3.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 3.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 0.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.9 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 4.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 6.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 3.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |