Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
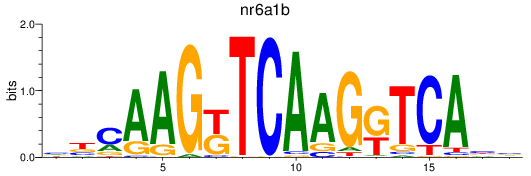
Results for nr6a1b
Z-value: 0.65
Transcription factors associated with nr6a1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr6a1b
|
ENSDARG00000014480 | nuclear receptor subfamily 6, group A, member 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr6a1b | dr11_v1_chr21_+_8198652_8198652 | 0.30 | 3.1e-03 | Click! |
Activity profile of nr6a1b motif
Sorted Z-values of nr6a1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_10539293 | 5.51 |
ENSDART00000182265
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr12_+_33395748 | 5.19 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr16_-_45235947 | 5.06 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr5_+_20693724 | 4.73 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr3_+_39540014 | 4.35 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr3_+_12652244 | 3.96 |
ENSDART00000165980
|
abca3b
|
ATP-binding cassette, sub-family A (ABC1), member 3b |
| chr5_-_50992690 | 3.90 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr3_-_40976463 | 3.43 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr23_-_19953089 | 3.39 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr16_+_23811554 | 3.27 |
ENSDART00000114336
|
si:dkey-7f3.9
|
si:dkey-7f3.9 |
| chr3_+_19299309 | 3.26 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr9_+_42270043 | 3.05 |
ENSDART00000137435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr23_-_10175898 | 3.03 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr4_-_17409533 | 3.00 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr3_-_40976288 | 2.80 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr12_-_18408566 | 2.66 |
ENSDART00000078767
ENSDART00000152748 |
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr23_-_24450686 | 2.47 |
ENSDART00000189161
|
spen
|
spen family transcriptional repressor |
| chr10_+_29260096 | 2.44 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr20_+_4392687 | 2.32 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr17_+_18117029 | 2.21 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr11_+_15878343 | 2.16 |
ENSDART00000167191
ENSDART00000171862 ENSDART00000163992 ENSDART00000170065 |
pank4
|
pantothenate kinase 4 |
| chr11_-_5563498 | 2.10 |
ENSDART00000160835
|
pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr16_+_52751445 | 2.09 |
ENSDART00000191807
|
atp6v1c1a
|
ATPase H+ transporting V1 subunit C1a |
| chr14_-_25095808 | 2.08 |
ENSDART00000184244
|
matr3l1.1
|
matrin 3-like 1.1 |
| chr6_+_39836474 | 2.07 |
ENSDART00000112637
|
smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr20_+_9128829 | 2.04 |
ENSDART00000064144
ENSDART00000137450 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr12_-_17024079 | 2.03 |
ENSDART00000129027
|
ifit11
|
interferon-induced protein with tetratricopeptide repeats 11 |
| chr2_+_24700922 | 1.88 |
ENSDART00000170467
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr24_-_25256230 | 1.84 |
ENSDART00000155780
|
hhla2b.1
|
HERV-H LTR-associating 2b, tandem duplicate 1 |
| chr4_-_63166758 | 1.80 |
ENSDART00000192765
|
CR450780.1
|
|
| chr5_+_50879545 | 1.79 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr7_+_19482084 | 1.79 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr8_-_2153147 | 1.75 |
ENSDART00000124093
|
si:dkeyp-117b11.1
|
si:dkeyp-117b11.1 |
| chr16_+_14216581 | 1.71 |
ENSDART00000113093
|
gba
|
glucosidase, beta, acid |
| chr3_+_1749793 | 1.62 |
ENSDART00000149308
|
si:dkeyp-52c3.7
|
si:dkeyp-52c3.7 |
| chr25_-_29415369 | 1.60 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr6_-_30932078 | 1.57 |
ENSDART00000028612
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr15_+_16886196 | 1.55 |
ENSDART00000139296
ENSDART00000049196 |
gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr5_+_1079423 | 1.55 |
ENSDART00000172231
|
si:zfos-128g4.2
|
si:zfos-128g4.2 |
| chr6_+_48618512 | 1.52 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr25_+_7492663 | 1.51 |
ENSDART00000166496
|
cat
|
catalase |
| chr17_-_40956035 | 1.48 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr17_-_39786222 | 1.43 |
ENSDART00000154515
|
pimr62
|
Pim proto-oncogene, serine/threonine kinase, related 62 |
| chr17_-_37054959 | 1.43 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr19_+_5135113 | 1.42 |
ENSDART00000151310
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr22_+_24603930 | 1.41 |
ENSDART00000180240
ENSDART00000164256 |
CU655842.1
|
|
| chr8_-_14091886 | 1.35 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr10_+_25219728 | 1.34 |
ENSDART00000193829
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr22_+_10215558 | 1.33 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr7_+_35268054 | 1.32 |
ENSDART00000113842
|
dpep2
|
dipeptidase 2 |
| chr4_+_76973771 | 1.32 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr13_-_21688176 | 1.29 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr21_-_32374656 | 1.28 |
ENSDART00000112550
|
mapk9
|
mitogen-activated protein kinase 9 |
| chr21_+_11401247 | 1.27 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr14_-_5678457 | 1.24 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr19_+_7878122 | 1.23 |
ENSDART00000131917
|
si:dkeyp-85e10.2
|
si:dkeyp-85e10.2 |
| chr21_-_43040780 | 1.18 |
ENSDART00000187037
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr19_+_5134624 | 1.18 |
ENSDART00000151324
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr17_+_18117358 | 1.18 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr8_+_22472584 | 1.18 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr19_+_30662529 | 1.16 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr4_+_69712223 | 1.11 |
ENSDART00000163035
ENSDART00000164475 |
si:dkey-238o14.1
zgc:174944
|
si:dkey-238o14.1 zgc:174944 |
| chr23_-_545942 | 1.08 |
ENSDART00000105316
|
samhd1
|
SAM domain and HD domain 1 |
| chr3_+_22442445 | 1.08 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr10_-_22057001 | 1.05 |
ENSDART00000016575
|
tlx3b
|
T cell leukemia homeobox 3b |
| chr25_-_225964 | 1.03 |
ENSDART00000193424
|
CABZ01113818.1
|
|
| chr6_+_41099787 | 1.00 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr12_+_46605600 | 0.99 |
ENSDART00000123533
|
FADS6
|
fatty acid desaturase 6 |
| chr6_+_39506043 | 0.96 |
ENSDART00000086260
|
CR388364.1
|
|
| chr15_-_41720173 | 0.96 |
ENSDART00000155585
ENSDART00000075909 |
ftr76
|
finTRIM family, member 76 |
| chr22_-_17781213 | 0.95 |
ENSDART00000137984
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr10_-_17284055 | 0.94 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr15_+_11840311 | 0.94 |
ENSDART00000167671
|
prkd2
|
protein kinase D2 |
| chr12_-_30583668 | 0.93 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr1_+_17527342 | 0.90 |
ENSDART00000139702
ENSDART00000140076 ENSDART00000005593 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr1_+_17527931 | 0.89 |
ENSDART00000131927
|
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr7_-_15370042 | 0.85 |
ENSDART00000128183
|
ANPEP
|
si:ch211-276a23.5 |
| chr25_+_33939728 | 0.84 |
ENSDART00000148537
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr6_+_2030703 | 0.84 |
ENSDART00000109679
ENSDART00000187502 ENSDART00000191165 ENSDART00000187544 |
quo
|
quattro |
| chr8_+_25145464 | 0.83 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr3_+_13469704 | 0.83 |
ENSDART00000166806
|
zgc:77748
|
zgc:77748 |
| chr4_+_9836465 | 0.82 |
ENSDART00000004879
|
hsp90b1
|
heat shock protein 90, beta (grp94), member 1 |
| chr19_+_20724347 | 0.79 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr4_-_49582758 | 0.79 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr3_-_7656059 | 0.78 |
ENSDART00000170917
|
junbb
|
JunB proto-oncogene, AP-1 transcription factor subunit b |
| chr6_+_40671336 | 0.76 |
ENSDART00000111639
ENSDART00000186617 |
rereb
|
arginine-glutamic acid dipeptide (RE) repeats b |
| chr3_+_32112004 | 0.74 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr4_-_71267461 | 0.70 |
ENSDART00000161219
ENSDART00000151838 |
si:dkeyp-51g9.4
|
si:dkeyp-51g9.4 |
| chr2_+_10771787 | 0.68 |
ENSDART00000187782
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr25_+_10830269 | 0.59 |
ENSDART00000175736
|
si:ch211-147g22.5
|
si:ch211-147g22.5 |
| chr20_+_13175379 | 0.57 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr4_+_7827261 | 0.57 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr15_+_44133972 | 0.54 |
ENSDART00000157846
ENSDART00000191082 |
CU655961.2
|
|
| chr13_+_8306399 | 0.54 |
ENSDART00000080339
|
galm
|
galactose mutarotase |
| chr16_-_46558521 | 0.54 |
ENSDART00000189406
|
si:dkey-152b24.8
|
si:dkey-152b24.8 |
| chr4_-_71470455 | 0.53 |
ENSDART00000172685
|
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr7_-_30639385 | 0.52 |
ENSDART00000173618
|
myo1ea
|
myosin IE, a |
| chr8_-_23578660 | 0.52 |
ENSDART00000039080
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr4_-_34776147 | 0.50 |
ENSDART00000167498
|
si:dkey-146m20.14
|
si:dkey-146m20.14 |
| chr4_-_47096170 | 0.47 |
ENSDART00000161079
|
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr25_-_210730 | 0.45 |
ENSDART00000187580
|
FP236318.1
|
|
| chr4_+_9478500 | 0.43 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr23_+_27779452 | 0.41 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr9_-_7287375 | 0.35 |
ENSDART00000128352
|
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr9_-_27749936 | 0.33 |
ENSDART00000064156
|
tbccd1
|
TBCC domain containing 1 |
| chr5_-_3839285 | 0.32 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr15_+_22722684 | 0.32 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr3_-_39627412 | 0.32 |
ENSDART00000123292
|
si:dkey-27o4.1
|
si:dkey-27o4.1 |
| chr4_-_38829098 | 0.30 |
ENSDART00000151932
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr1_+_57348756 | 0.29 |
ENSDART00000063750
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr16_-_46558708 | 0.24 |
ENSDART00000128037
|
si:dkey-152b24.8
|
si:dkey-152b24.8 |
| chr4_-_70263609 | 0.17 |
ENSDART00000163868
|
si:dkey-3h2.3
|
si:dkey-3h2.3 |
| chr8_-_54216499 | 0.16 |
ENSDART00000193678
|
mbd4
|
methyl-CpG binding domain protein 4 |
| chr23_+_23119008 | 0.15 |
ENSDART00000132418
|
samd11
|
sterile alpha motif domain containing 11 |
| chr1_-_49947290 | 0.14 |
ENSDART00000141476
|
sgms2
|
sphingomyelin synthase 2 |
| chr1_-_8566567 | 0.12 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr2_+_13694450 | 0.10 |
ENSDART00000077259
ENSDART00000189485 |
ebna1bp2
|
EBNA1 binding protein 2 |
| chr4_-_47095583 | 0.09 |
ENSDART00000183595
ENSDART00000183279 ENSDART00000191638 |
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr24_+_19542323 | 0.08 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr12_+_30788912 | 0.06 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr16_+_27224068 | 0.04 |
ENSDART00000059013
|
sec61b
|
Sec61 translocon beta subunit |
| chr23_+_17102960 | 0.02 |
ENSDART00000053414
|
commd7
|
COMM domain containing 7 |
| chr4_-_63559311 | 0.01 |
ENSDART00000164415
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr6a1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 1.4 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.6 | 1.8 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.6 | 1.8 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.6 | 1.7 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.5 | 3.3 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.4 | 1.3 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.4 | 1.6 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.4 | 1.1 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 2.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.3 | 6.1 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.3 | 0.8 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.3 | 3.0 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 1.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.9 | GO:0003272 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.1 | 1.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.8 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 1.3 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 2.0 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 1.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.3 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 5.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.4 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 1.4 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.9 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 3.8 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 2.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.6 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 2.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.9 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 0.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.3 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 3.4 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.5 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.8 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 1.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 5.4 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.3 | 2.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.3 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 2.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 2.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 3.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 2.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.5 | GO:0005902 | microvillus(GO:0005902) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.5 | 1.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 1.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.4 | 2.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 3.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 1.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.2 | 1.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 1.1 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.2 | 1.5 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 2.7 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 1.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 3.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 2.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 6.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 2.0 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 2.1 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.1 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 3.9 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 1.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 5.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 4.0 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.6 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 2.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 4.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.7 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 1.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 3.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.5 | 2.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 1.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 1.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |