Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
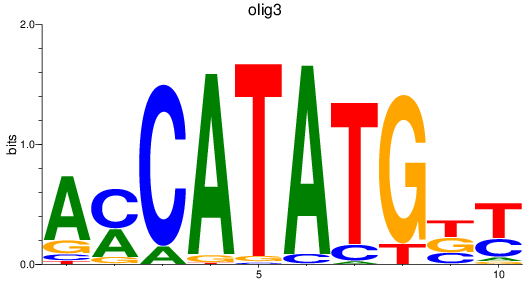
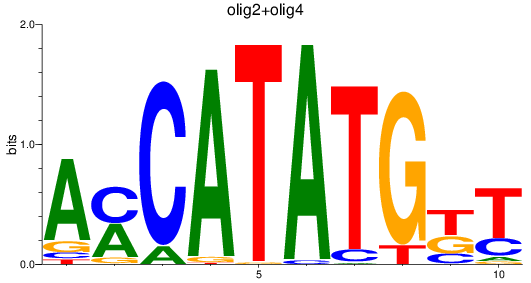
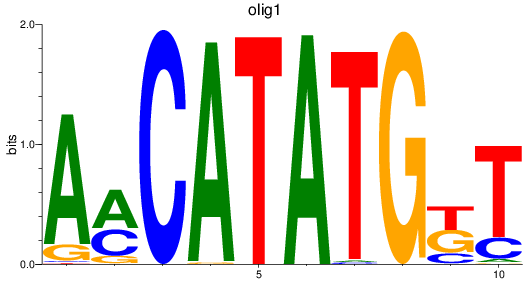
Results for olig3_olig2+olig4_olig1
Z-value: 1.05
Transcription factors associated with olig3_olig2+olig4_olig1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
olig3
|
ENSDARG00000074253 | oligodendrocyte transcription factor 3 |
|
olig2
|
ENSDARG00000040946 | oligodendrocyte lineage transcription factor 2 |
|
olig4
|
ENSDARG00000052610 | oligodendrocyte transcription factor 4 |
|
olig4
|
ENSDARG00000116678 | oligodendrocyte transcription factor 4 |
|
olig1
|
ENSDARG00000040948 | oligodendrocyte transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| olig2 | dr11_v1_chr9_-_32753535_32753535 | 0.57 | 2.6e-09 | Click! |
| olig1 | dr11_v1_chr9_-_32730487_32730487 | 0.55 | 8.2e-09 | Click! |
| olig4 | dr11_v1_chr13_+_45967179_45967179 | 0.46 | 2.6e-06 | Click! |
Activity profile of olig3_olig2+olig4_olig1 motif
Sorted Z-values of olig3_olig2+olig4_olig1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_22012389 | 8.20 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr4_-_8882572 | 7.77 |
ENSDART00000190060
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr7_-_38633867 | 6.86 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr23_+_26491931 | 6.84 |
ENSDART00000190124
ENSDART00000132905 |
si:ch73-206d17.1
|
si:ch73-206d17.1 |
| chr23_-_11870962 | 6.72 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr14_+_25817246 | 6.70 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr14_-_25599002 | 6.70 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr2_-_1518360 | 6.65 |
ENSDART00000182788
|
c8b
|
complement component 8, beta polypeptide |
| chr13_+_11439486 | 6.50 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr12_-_14922955 | 6.23 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr15_+_22267847 | 6.23 |
ENSDART00000110665
|
spa17
|
sperm autoantigenic protein 17 |
| chr23_-_21453614 | 6.13 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr14_-_41285392 | 5.76 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr2_-_36040820 | 5.74 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr16_+_23972126 | 5.68 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr2_-_39036604 | 5.68 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr2_-_27775236 | 5.64 |
ENSDART00000187983
|
XKR4
|
zgc:123035 |
| chr5_-_27994679 | 5.40 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr21_+_11468642 | 5.21 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr23_+_21544227 | 5.03 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr15_-_21165237 | 5.03 |
ENSDART00000157069
|
A2ML1 (1 of many)
|
si:ch211-212c13.8 |
| chr8_-_54223316 | 4.81 |
ENSDART00000018054
|
trh
|
thyrotropin-releasing hormone |
| chr23_+_4299887 | 4.76 |
ENSDART00000132604
|
l3mbtl1a
|
l(3)mbt-like 1a (Drosophila) |
| chr20_+_34913069 | 4.71 |
ENSDART00000007584
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr14_-_4682114 | 4.70 |
ENSDART00000014454
|
gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr21_+_6751405 | 4.68 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr18_+_30878196 | 4.67 |
ENSDART00000099326
ENSDART00000146041 |
mthfsd
|
methenyltetrahydrofolate synthetase domain containing |
| chr18_+_13164325 | 4.64 |
ENSDART00000189057
|
tat
|
tyrosine aminotransferase |
| chr1_-_16507812 | 4.60 |
ENSDART00000169081
|
mtmr7b
|
myotubularin related protein 7b |
| chr23_+_20563779 | 4.60 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr7_-_31938938 | 4.30 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
| chr15_+_4988189 | 4.27 |
ENSDART00000142995
ENSDART00000062852 |
spcs2
|
signal peptidase complex subunit 2 |
| chr25_+_3994823 | 4.17 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr25_-_26753196 | 4.12 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr2_+_24177006 | 4.02 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr6_+_48618512 | 3.92 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr12_-_10220036 | 3.86 |
ENSDART00000134619
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr25_-_5740334 | 3.83 |
ENSDART00000169622
ENSDART00000168720 |
LO017739.1
|
|
| chr5_+_52625975 | 3.80 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr13_+_40019001 | 3.75 |
ENSDART00000158820
|
golga7bb
|
golgin A7 family, member Bb |
| chr14_+_31618982 | 3.71 |
ENSDART00000026195
|
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr13_+_23677949 | 3.68 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr25_-_11057753 | 3.62 |
ENSDART00000186551
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr21_+_31150438 | 3.60 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr5_-_35301800 | 3.60 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr11_+_36158134 | 3.58 |
ENSDART00000189827
ENSDART00000163330 |
grm2b
|
glutamate receptor, metabotropic 2b |
| chr13_-_24311628 | 3.58 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr4_+_15968483 | 3.55 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr21_+_11415224 | 3.46 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr24_-_9689915 | 3.44 |
ENSDART00000185972
ENSDART00000093046 |
uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr20_+_25486021 | 3.43 |
ENSDART00000063052
|
hook1
|
hook microtubule-tethering protein 1 |
| chr6_-_13308813 | 3.42 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr6_+_27151940 | 3.41 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr25_+_37397031 | 3.39 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr16_+_50289916 | 3.31 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr7_+_1306723 | 3.30 |
ENSDART00000189868
|
si:dkeyp-74b6.2
|
si:dkeyp-74b6.2 |
| chr17_+_15534815 | 3.27 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr21_+_53504 | 3.24 |
ENSDART00000170452
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr22_+_4707663 | 3.24 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr2_-_44282796 | 3.24 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr7_+_39634873 | 3.23 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr13_+_771403 | 3.15 |
ENSDART00000093166
|
nrxn1b
|
neurexin 1b |
| chr1_-_22803147 | 3.14 |
ENSDART00000086867
|
tapt1b
|
transmembrane anterior posterior transformation 1b |
| chr18_+_38192499 | 3.11 |
ENSDART00000191849
|
nucb2b
|
nucleobindin 2b |
| chr3_+_43374571 | 3.01 |
ENSDART00000182497
|
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr16_-_8132742 | 2.99 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr14_+_35405518 | 2.98 |
ENSDART00000171565
|
zbtb3
|
zinc finger and BTB domain containing 3 |
| chr21_+_31150773 | 2.96 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr9_+_38168012 | 2.95 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr15_+_15856178 | 2.88 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr10_-_26226228 | 2.86 |
ENSDART00000182967
|
fhdc3
|
FH2 domain containing 3 |
| chr14_-_47314011 | 2.85 |
ENSDART00000178523
|
fstl5
|
follistatin-like 5 |
| chr24_-_21172122 | 2.85 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr18_+_6866276 | 2.85 |
ENSDART00000187516
|
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr8_-_37391372 | 2.81 |
ENSDART00000190407
ENSDART00000009569 |
slc12a5b
|
solute carrier family 12 (potassium/chloride transporter), member 5b |
| chr18_-_40708537 | 2.80 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr7_-_18598661 | 2.79 |
ENSDART00000182109
|
DTX4
|
si:ch211-119e14.2 |
| chr19_-_7406933 | 2.78 |
ENSDART00000151137
|
oxr1b
|
oxidation resistance 1b |
| chr1_+_29759678 | 2.75 |
ENSDART00000054059
ENSDART00000101856 |
cpb2
|
carboxypeptidase B2 (plasma) |
| chr15_+_36115955 | 2.74 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr21_-_26918901 | 2.73 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr5_+_20148671 | 2.73 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr17_-_42218652 | 2.72 |
ENSDART00000081396
ENSDART00000190007 |
nkx2.2a
|
NK2 homeobox 2a |
| chr2_-_39017838 | 2.72 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr16_+_5156420 | 2.70 |
ENSDART00000012053
|
elovl4a
|
ELOVL fatty acid elongase 4a |
| chr1_-_22652424 | 2.66 |
ENSDART00000036797
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr17_-_43594864 | 2.66 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr19_-_8798178 | 2.63 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr10_-_4961923 | 2.61 |
ENSDART00000050177
ENSDART00000146066 |
snx30
|
sorting nexin family member 30 |
| chr21_-_2310064 | 2.58 |
ENSDART00000169520
|
si:ch211-241b2.1
|
si:ch211-241b2.1 |
| chr13_-_9525527 | 2.58 |
ENSDART00000190618
|
CR848040.5
|
|
| chr7_+_72630369 | 2.58 |
ENSDART00000170698
|
FO905040.1
|
|
| chr22_+_15979430 | 2.57 |
ENSDART00000189703
ENSDART00000192674 |
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr19_-_24125457 | 2.52 |
ENSDART00000080632
|
zgc:64022
|
zgc:64022 |
| chr19_+_9277327 | 2.52 |
ENSDART00000104623
ENSDART00000151164 |
si:rp71-15k1.1
|
si:rp71-15k1.1 |
| chr6_+_54142311 | 2.51 |
ENSDART00000154115
|
hmga1b
|
high mobility group AT-hook 1b |
| chr20_+_52529044 | 2.51 |
ENSDART00000158230
ENSDART00000002787 |
pycr3
|
pyrroline-5-carboxylate reductase 3 |
| chr13_+_15581270 | 2.50 |
ENSDART00000189880
ENSDART00000190067 ENSDART00000041293 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr2_-_5074812 | 2.47 |
ENSDART00000163728
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr18_-_30020879 | 2.44 |
ENSDART00000162086
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr14_+_25816874 | 2.43 |
ENSDART00000005499
|
glra1
|
glycine receptor, alpha 1 |
| chr9_+_22929675 | 2.43 |
ENSDART00000061299
|
tsn
|
translin |
| chr6_-_609880 | 2.43 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr2_+_24177190 | 2.42 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr4_-_4507761 | 2.39 |
ENSDART00000130588
|
tbc1d30
|
TBC1 domain family, member 30 |
| chr7_-_24022340 | 2.39 |
ENSDART00000149133
|
cideb
|
cell death-inducing DFFA-like effector b |
| chr10_+_5689510 | 2.38 |
ENSDART00000183217
ENSDART00000172632 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_7147226 | 2.37 |
ENSDART00000178155
|
BX005085.5
|
|
| chr2_-_17947389 | 2.36 |
ENSDART00000190089
ENSDART00000191872 ENSDART00000184039 ENSDART00000179791 |
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr22_+_38194151 | 2.35 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr8_+_39998467 | 2.35 |
ENSDART00000073782
ENSDART00000134452 |
ggt5a
|
gamma-glutamyltransferase 5a |
| chr3_-_30625219 | 2.35 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr13_+_18202703 | 2.33 |
ENSDART00000109642
|
tet1
|
tet methylcytosine dioxygenase 1 |
| chr3_-_52674089 | 2.31 |
ENSDART00000154260
ENSDART00000125455 |
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr16_+_23984755 | 2.31 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr10_-_23358357 | 2.31 |
ENSDART00000135475
|
cadm2a
|
cell adhesion molecule 2a |
| chr22_-_16034137 | 2.31 |
ENSDART00000062629
|
zbtb37
|
zinc finger and BTB domain containing 37 |
| chr20_-_34028967 | 2.30 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr24_-_30096666 | 2.29 |
ENSDART00000183285
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr25_-_37338048 | 2.29 |
ENSDART00000073439
|
trim44
|
tripartite motif containing 44 |
| chr16_-_44399335 | 2.28 |
ENSDART00000165058
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr17_-_36896560 | 2.28 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr25_+_19954576 | 2.26 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr3_-_57744323 | 2.26 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr6_+_58492201 | 2.23 |
ENSDART00000156375
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr9_+_42066030 | 2.22 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr17_-_8692722 | 2.21 |
ENSDART00000148931
ENSDART00000192891 |
ctbp2a
|
C-terminal binding protein 2a |
| chr17_-_12389259 | 2.20 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr17_-_23727978 | 2.20 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr14_+_36521553 | 2.19 |
ENSDART00000136233
|
TENM3
|
si:dkey-237h12.3 |
| chr12_+_3912544 | 2.19 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr9_+_42095220 | 2.18 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr15_-_25153352 | 2.16 |
ENSDART00000078095
ENSDART00000122184 |
vps53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr21_+_34849406 | 2.15 |
ENSDART00000065325
|
cdc23
|
CDC23 (cell division cycle 23, yeast, homolog) |
| chr24_-_10828560 | 2.14 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr18_+_50278858 | 2.13 |
ENSDART00000014582
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr17_-_37184655 | 2.11 |
ENSDART00000180447
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr21_-_22114625 | 2.10 |
ENSDART00000177426
ENSDART00000135410 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr25_+_7423770 | 2.07 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr2_+_50608099 | 2.06 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr3_+_40856095 | 2.06 |
ENSDART00000143207
|
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr23_+_7548797 | 2.06 |
ENSDART00000006765
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr10_-_26225548 | 2.06 |
ENSDART00000132019
ENSDART00000079194 |
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr11_+_44502410 | 2.02 |
ENSDART00000172998
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr2_-_22966076 | 2.02 |
ENSDART00000143412
ENSDART00000146014 ENSDART00000183443 ENSDART00000191056 ENSDART00000183539 |
sap130b
|
Sin3A-associated protein b |
| chr6_+_49583153 | 2.01 |
ENSDART00000103357
|
vapb
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr7_-_71434298 | 2.00 |
ENSDART00000180507
|
lgi2a
|
leucine-rich repeat LGI family, member 2a |
| chr8_+_6863462 | 1.99 |
ENSDART00000064163
|
nefmb
|
neurofilament, medium polypeptide b |
| chr7_-_49654492 | 1.98 |
ENSDART00000174324
|
hrasb
|
-Ha-ras Harvey rat sarcoma viral oncogene homolog b |
| chr5_-_16983336 | 1.98 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr12_+_24562667 | 1.97 |
ENSDART00000056256
|
nrxn1a
|
neurexin 1a |
| chr3_-_34801041 | 1.96 |
ENSDART00000103043
|
nsfa
|
N-ethylmaleimide-sensitive factor a |
| chr21_+_22404662 | 1.93 |
ENSDART00000183455
|
lmbrd2b
|
LMBR1 domain containing 2b |
| chr19_+_9113932 | 1.92 |
ENSDART00000060442
|
setdb1a
|
SET domain, bifurcated 1a |
| chr21_-_22115136 | 1.90 |
ENSDART00000134715
ENSDART00000089246 ENSDART00000139789 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr4_+_4272622 | 1.88 |
ENSDART00000164810
|
ano2
|
anoctamin 2 |
| chr1_+_10720294 | 1.86 |
ENSDART00000139387
|
atp1b1b
|
ATPase Na+/K+ transporting subunit beta 1b |
| chr2_-_7189594 | 1.85 |
ENSDART00000139703
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr19_+_38167468 | 1.83 |
ENSDART00000160756
|
phf14
|
PHD finger protein 14 |
| chr9_+_32859967 | 1.83 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr12_-_17863467 | 1.83 |
ENSDART00000042006
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr5_-_72178739 | 1.82 |
ENSDART00000050971
|
rab14l
|
RAB14, member RAS oncogene family, like |
| chr16_-_31452416 | 1.82 |
ENSDART00000140880
ENSDART00000008297 ENSDART00000147373 |
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr19_+_25649626 | 1.82 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr20_-_28800999 | 1.81 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr2_+_55429988 | 1.81 |
ENSDART00000166920
|
ap1m1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr18_-_39702327 | 1.80 |
ENSDART00000149158
|
dmxl2
|
Dmx-like 2 |
| chr17_+_24747209 | 1.79 |
ENSDART00000154623
|
stac
|
SH3 and cysteine rich domain |
| chr2_-_45154550 | 1.79 |
ENSDART00000148595
ENSDART00000150002 |
capn10
|
calpain 10 |
| chr19_+_12801940 | 1.79 |
ENSDART00000040073
|
mc5ra
|
melanocortin 5a receptor |
| chr6_-_50730749 | 1.78 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr1_-_21538673 | 1.77 |
ENSDART00000131257
|
wdr19
|
WD repeat domain 19 |
| chr1_-_29758947 | 1.77 |
ENSDART00000049514
ENSDART00000183571 ENSDART00000140345 |
alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr19_+_41006975 | 1.77 |
ENSDART00000138555
ENSDART00000049842 |
casd1
|
CAS1 domain containing 1 |
| chr1_-_17711636 | 1.76 |
ENSDART00000148322
ENSDART00000122670 |
ufsp2
|
ufm1-specific peptidase 2 |
| chr22_+_3238474 | 1.76 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr3_+_43774369 | 1.75 |
ENSDART00000157964
|
zc3h7a
|
zinc finger CCCH-type containing 7A |
| chr19_+_17259912 | 1.75 |
ENSDART00000078951
|
MANEAL
|
si:ch211-30b16.2 |
| chr1_-_17693273 | 1.74 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr17_+_6956696 | 1.74 |
ENSDART00000171368
|
zgc:172341
|
zgc:172341 |
| chr23_-_18057270 | 1.74 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr24_-_27400630 | 1.72 |
ENSDART00000165760
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr25_+_17339814 | 1.71 |
ENSDART00000141311
|
cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr19_+_11984725 | 1.71 |
ENSDART00000185960
|
spag1a
|
sperm associated antigen 1a |
| chr1_+_40613297 | 1.71 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr23_+_23658474 | 1.70 |
ENSDART00000162838
|
agrn
|
agrin |
| chr20_-_46467280 | 1.70 |
ENSDART00000060702
|
rmdn3
|
regulator of microtubule dynamics 3 |
| chr20_+_23670494 | 1.69 |
ENSDART00000155262
|
nek1
|
NIMA-related kinase 1 |
| chr23_+_17522867 | 1.69 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr19_-_46037835 | 1.69 |
ENSDART00000163815
|
nup153
|
nucleoporin 153 |
| chr8_-_12468744 | 1.68 |
ENSDART00000135019
|
FIBCD1 (1 of many)
|
si:dkeyp-51b7.3 |
| chr16_-_26437668 | 1.67 |
ENSDART00000142056
|
megf8
|
multiple EGF-like-domains 8 |
| chr2_+_54755172 | 1.66 |
ENSDART00000097864
|
ankrd12
|
ankyrin repeat domain 12 |
| chr16_-_39267185 | 1.66 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr16_-_8342749 | 1.66 |
ENSDART00000028108
|
ddc
|
dopa decarboxylase |
| chr16_+_25074029 | 1.66 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr25_+_21833287 | 1.64 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr14_-_2327825 | 1.63 |
ENSDART00000180328
ENSDART00000191135 ENSDART00000114302 ENSDART00000189869 |
pcdh2ab8
pcdh2aa1
|
protocadherin 2 alpha b 8 protocadherin 2 alpha a 1 |
| chr20_-_3997531 | 1.62 |
ENSDART00000092217
|
ttc13
|
tetratricopeptide repeat domain 13 |
| chr5_-_24245218 | 1.62 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
Network of associatons between targets according to the STRING database.
First level regulatory network of olig3_olig2+olig4_olig1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 1.9 | 5.7 | GO:0032369 | negative regulation of lipid transport(GO:0032369) negative regulation of lipid catabolic process(GO:0050995) |
| 1.6 | 4.8 | GO:0001692 | histamine metabolic process(GO:0001692) negative regulation of anion transport(GO:1903792) |
| 1.6 | 6.3 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 1.6 | 6.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 1.0 | 3.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.9 | 2.8 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.9 | 2.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.9 | 2.7 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.8 | 3.2 | GO:0035521 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.8 | 4.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.7 | 4.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.7 | 3.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.6 | 1.8 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.6 | 1.7 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.5 | 2.6 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.5 | 2.0 | GO:0048940 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.5 | 1.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.5 | 3.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.5 | 2.4 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.5 | 1.4 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.5 | 2.3 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.5 | 1.8 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.4 | 1.3 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.4 | 1.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.4 | 2.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.4 | 1.3 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.4 | 5.7 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.4 | 1.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.4 | 3.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.4 | 5.7 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.4 | 2.4 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.4 | 1.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.4 | 1.6 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.4 | 1.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 1.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.4 | 4.2 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 1.1 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.4 | 1.5 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.4 | 1.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.4 | 1.8 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.4 | 2.1 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.4 | 1.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.4 | 2.5 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.4 | 1.4 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.3 | 1.0 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.3 | 3.0 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.3 | 2.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 2.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 | 5.9 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.3 | 3.6 | GO:0001840 | neural plate development(GO:0001840) |
| 0.3 | 1.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.3 | 2.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.3 | 1.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.3 | 0.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.3 | 3.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.3 | 3.3 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.3 | 1.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.3 | 1.8 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 5.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 1.1 | GO:0014060 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.3 | 2.5 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.7 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 2.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 2.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.3 | 2.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 1.5 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.2 | 0.7 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 7.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 1.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 1.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 1.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 0.7 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 0.5 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 0.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 3.5 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.2 | 4.7 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.2 | 0.7 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.2 | 0.7 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.2 | 3.1 | GO:0032094 | response to food(GO:0032094) |
| 0.2 | 5.8 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.2 | 1.3 | GO:0050951 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.2 | 1.3 | GO:1901490 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 0.2 | 3.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 1.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.2 | 2.0 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 0.8 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.2 | 0.6 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.2 | 1.0 | GO:0070814 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.2 | 6.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.2 | 2.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 0.9 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.2 | 4.3 | GO:0071542 | multicellular organismal response to stress(GO:0033555) dopaminergic neuron differentiation(GO:0071542) |
| 0.2 | 0.4 | GO:1900193 | regulation of oocyte maturation(GO:1900193) |
| 0.2 | 2.2 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 1.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 5.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 0.9 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.2 | 1.4 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.2 | 0.5 | GO:0044108 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.2 | 2.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 0.5 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 0.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.2 | 1.2 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.2 | 2.8 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 0.5 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.2 | 0.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.6 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 10.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.4 | GO:0002714 | positive regulation of B cell mediated immunity(GO:0002714) positive regulation of immunoglobulin mediated immune response(GO:0002891) histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.4 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.1 | 3.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 2.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.6 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 0.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.5 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 1.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 1.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.5 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 2.7 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.5 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 1.4 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 1.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 2.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 2.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 2.0 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.1 | 4.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 2.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 7.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.2 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 1.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.4 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.1 | 0.4 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 4.7 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.1 | 1.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 3.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 2.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 1.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.6 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.2 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.1 | 0.7 | GO:1901673 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 2.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.8 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 3.7 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.6 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.9 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 4.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.3 | GO:0000256 | allantoin catabolic process(GO:0000256) |
| 0.1 | 2.9 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.1 | 2.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.6 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.6 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 4.4 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.1 | 0.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 2.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 1.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.6 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.1 | 1.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.9 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.2 | GO:1902103 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 2.3 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 1.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.0 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 2.2 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.1 | 0.9 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.3 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 1.5 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 | 1.7 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.5 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.7 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 0.2 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 1.0 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.1 | 1.2 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 5.4 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.4 | GO:1902023 | ornithine transport(GO:0015822) L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-ornithine transmembrane transport(GO:1903352) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.2 | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902229) |
| 0.1 | 2.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 0.6 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.6 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 1.9 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.1 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 1.0 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.5 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.1 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.5 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.2 | GO:0006266 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 4.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.5 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.0 | 3.1 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 1.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.8 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 2.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.4 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 4.9 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.9 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.6 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 6.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.7 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 1.1 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.0 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 5.0 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.5 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.2 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 6.0 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 4.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.6 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.3 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.3 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.3 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.0 | GO:0016094 | polyprenol metabolic process(GO:0016093) polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 4.0 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 1.3 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.3 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.4 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.1 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.5 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.4 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.7 | GO:0050771 | negative regulation of axon extension(GO:0030517) negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.9 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.9 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.1 | 4.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 1.0 | 2.9 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.9 | 5.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.6 | 3.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 8.0 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.5 | 3.7 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.5 | 1.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.5 | 4.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.5 | 1.8 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.4 | 1.3 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.4 | 3.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 2.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.4 | 1.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.4 | 2.7 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.4 | 1.6 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.4 | 1.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.4 | 2.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.3 | 5.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 1.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 2.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 1.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 0.9 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.3 | 1.4 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.3 | 4.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.3 | 2.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 1.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 9.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 1.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.6 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.2 | 3.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 1.9 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 2.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 3.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 2.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 2.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 7.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.0 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 4.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 4.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.9 | GO:0031464 | inclusion body(GO:0016234) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 1.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.7 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 3.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 0.5 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.6 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 2.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 2.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 2.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 4.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 6.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 2.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.9 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.1 | 6.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 4.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 2.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 10.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 5.1 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 0.4 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 1.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 4.2 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 1.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.3 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 4.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 10.8 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 2.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.3 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 3.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 2.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.1 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 11.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.3 | GO:0031984 | organelle subcompartment(GO:0031984) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 13.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 2.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.4 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.0 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.5 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.4 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 1.0 | 9.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.9 | 5.4 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.8 | 6.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.8 | 3.2 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.7 | 2.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.6 | 4.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.6 | 1.8 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.6 | 1.8 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.6 | 4.0 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.6 | 2.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.5 | 2.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.5 | 2.7 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.5 | 1.6 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.5 | 3.6 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.5 | 2.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 2.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.5 | 1.5 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.5 | 7.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 1.3 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.4 | 5.7 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.4 | 2.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.4 | 4.6 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 1.6 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.4 | 3.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 1.5 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.3 | 4.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 2.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 2.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 5.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.3 | 1.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 2.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.3 | 1.7 | GO:0042974 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.3 | 0.8 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.3 | 1.4 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.3 | 2.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 2.4 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.3 | 5.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 2.5 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 1.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 1.0 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 2.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 0.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 0.6 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.2 | 3.9 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.2 | 0.6 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.2 | 1.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 1.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 0.8 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 0.8 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 1.0 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 1.8 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 1.0 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.2 | 1.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 1.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 4.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 6.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 0.7 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.2 | 2.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.7 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 0.5 | GO:0031073 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.2 | 4.6 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.2 | 1.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 4.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 3.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 2.0 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 0.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 1.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 2.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.0 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.7 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 2.9 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 1.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 3.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 2.7 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 6.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.7 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.2 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.1 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 2.8 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.4 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 1.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 2.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 3.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 2.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 2.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 4.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 1.3 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.8 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 2.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 4.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 6.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.7 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 9.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 2.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 4.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.3 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 1.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 2.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.6 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 0.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.7 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.6 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 1.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 3.3 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 2.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 2.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 1.3 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 4.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 2.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 3.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 14.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.8 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 10.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.5 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 10.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.5 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 4.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 2.2 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 3.1 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 3.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 3.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 3.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.8 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 25.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 4.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 1.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 4.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 3.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 1.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 1.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.6 | 14.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.4 | 4.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 2.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 1.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 8.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 1.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 4.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 3.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 0.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 5.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 1.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 2.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |