Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
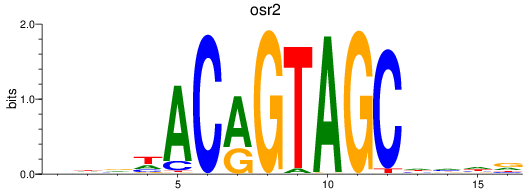
Results for osr2
Z-value: 0.45
Transcription factors associated with osr2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
osr2
|
ENSDARG00000038006 | odd-skipped related transciption factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| osr2 | dr11_v1_chr16_-_54405976_54405976 | -0.16 | 1.1e-01 | Click! |
Activity profile of osr2 motif
Sorted Z-values of osr2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_46010 | 7.01 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr18_-_45736 | 6.86 |
ENSDART00000148373
ENSDART00000148950 |
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr20_-_29418620 | 6.25 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr23_+_36095260 | 2.73 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr23_-_9925568 | 2.62 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr23_+_7379728 | 1.91 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr2_+_5446882 | 1.75 |
ENSDART00000083260
|
dusp28
|
dual specificity phosphatase 28 |
| chr8_-_4100365 | 1.40 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr13_-_27621246 | 1.29 |
ENSDART00000102232
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr22_-_5006801 | 1.27 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr7_+_17374560 | 1.24 |
ENSDART00000172272
ENSDART00000073467 |
nitr7a
|
novel immune-type receptor 7a |
| chr20_-_26866111 | 1.23 |
ENSDART00000077767
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr18_-_977075 | 1.15 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr2_-_48497175 | 1.13 |
ENSDART00000158839
|
CR391991.5
|
|
| chr3_-_30384353 | 1.11 |
ENSDART00000186832
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr2_-_48448225 | 1.11 |
ENSDART00000129826
|
CR391991.2
|
|
| chr5_+_6617401 | 1.05 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr10_+_21899753 | 1.05 |
ENSDART00000080155
|
hrh2b
|
histamine receptor H2b |
| chr7_+_42206543 | 1.04 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr21_+_21679086 | 0.97 |
ENSDART00000146225
|
or125-5
|
odorant receptor, family E, subfamily 125, member 5 |
| chr7_-_17771166 | 0.94 |
ENSDART00000173620
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr14_+_29945070 | 0.94 |
ENSDART00000185039
|
fam149a
|
family with sequence similarity 149 member A |
| chr14_+_5383060 | 0.94 |
ENSDART00000187825
|
lbx2
|
ladybird homeobox 2 |
| chr22_-_11626014 | 0.90 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr5_-_48307804 | 0.89 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr15_-_31357634 | 0.88 |
ENSDART00000127485
|
or111-2
|
odorant receptor, family D, subfamily 111, member 2 |
| chr6_+_19383267 | 0.80 |
ENSDART00000166549
|
mchr1a
|
melanin-concentrating hormone receptor 1a |
| chr5_-_61787969 | 0.77 |
ENSDART00000112744
|
gas2l2
|
growth arrest specific 2 like 2 |
| chr6_+_54576520 | 0.72 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr25_-_19395156 | 0.57 |
ENSDART00000155335
|
map1ab
|
microtubule-associated protein 1Ab |
| chr1_+_40199074 | 0.49 |
ENSDART00000179679
ENSDART00000136849 |
si:ch211-113e8.5
|
si:ch211-113e8.5 |
| chr8_+_48858132 | 0.38 |
ENSDART00000124737
ENSDART00000079644 |
tp73
|
tumor protein p73 |
| chr21_-_40834413 | 0.38 |
ENSDART00000148513
|
limk1b
|
LIM domain kinase 1b |
| chr2_-_48512450 | 0.37 |
ENSDART00000160132
|
CR391991.1
|
|
| chr12_+_28856151 | 0.36 |
ENSDART00000152969
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr13_+_29778610 | 0.35 |
ENSDART00000132004
|
pax2a
|
paired box 2a |
| chr1_+_44826593 | 0.34 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr3_-_10751491 | 0.29 |
ENSDART00000016351
|
zgc:112965
|
zgc:112965 |
| chr2_+_3986083 | 0.27 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr6_+_46431848 | 0.26 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr8_-_22639794 | 0.23 |
ENSDART00000188029
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr19_-_25519310 | 0.22 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr6_-_40884453 | 0.16 |
ENSDART00000017968
ENSDART00000154100 |
sirt4
|
sirtuin 4 |
| chr14_+_36246726 | 0.14 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr23_-_4704938 | 0.10 |
ENSDART00000067293
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr19_-_9503473 | 0.09 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr16_-_13612650 | 0.07 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr23_-_36003282 | 0.00 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of osr2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.9 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.7 | 2.7 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.5 | 1.9 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 6.2 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.2 | 0.9 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 2.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.9 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.9 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 1.5 | GO:0021782 | glial cell development(GO:0021782) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 13.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.4 | 13.9 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.3 | 0.8 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.2 | 0.9 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.8 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.0 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 13.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |