Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
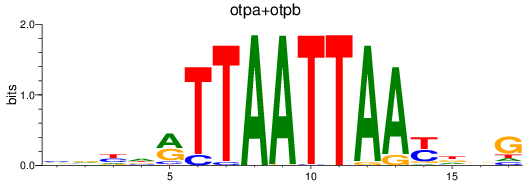
Results for otpa+otpb
Z-value: 0.69
Transcription factors associated with otpa+otpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
otpa
|
ENSDARG00000014201 | orthopedia homeobox a |
|
otpb
|
ENSDARG00000058379 | orthopedia homeobox b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otpb | dr11_v1_chr5_-_51619742_51619742 | 0.20 | 4.9e-02 | Click! |
| otpa | dr11_v1_chr21_+_7582036_7582059 | 0.06 | 5.6e-01 | Click! |
Activity profile of otpa+otpb motif
Sorted Z-values of otpa+otpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_31713239 | 11.60 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr5_+_2815021 | 9.61 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr17_+_15433518 | 8.77 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433671 | 8.65 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr10_+_26747755 | 8.14 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr9_+_34127005 | 7.27 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr15_-_21014270 | 5.65 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr2_-_17392799 | 4.95 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr11_+_40812590 | 4.75 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr2_-_17393216 | 4.51 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr6_+_612594 | 4.04 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr17_+_16564921 | 3.69 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr17_+_50524573 | 3.68 |
ENSDART00000187974
|
CR382385.1
|
|
| chr3_+_3681116 | 3.59 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr18_+_41527877 | 2.57 |
ENSDART00000146972
|
selenot1b
|
selenoprotein T, 1b |
| chr13_-_52089003 | 2.53 |
ENSDART00000187600
|
tmem254
|
transmembrane protein 254 |
| chr4_-_9891874 | 2.49 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr14_-_33481428 | 2.42 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr22_+_19552987 | 2.27 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr7_-_34448076 | 2.24 |
ENSDART00000170935
|
nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr22_-_21897203 | 2.12 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr9_+_48761455 | 2.06 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr10_-_27049170 | 2.03 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr4_-_1801519 | 2.03 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr25_+_21833287 | 2.00 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr16_+_40024883 | 1.94 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr2_+_50608099 | 1.82 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr21_-_20939488 | 1.77 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr21_-_2814709 | 1.73 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr8_-_44926077 | 1.73 |
ENSDART00000084417
|
timm17b
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr14_-_48765262 | 1.72 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr5_+_67390115 | 1.60 |
ENSDART00000193255
|
ebf2
|
early B cell factor 2 |
| chr9_+_2574122 | 1.59 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr6_-_11768198 | 1.59 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr12_-_35830625 | 1.53 |
ENSDART00000180028
|
CU459056.1
|
|
| chr22_-_16154771 | 1.52 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr4_+_9669717 | 1.51 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr21_+_22840246 | 1.50 |
ENSDART00000151621
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr7_-_52388734 | 1.48 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr15_+_5360407 | 1.45 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr20_+_25586099 | 1.40 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr11_-_1509773 | 1.34 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr16_+_46111849 | 1.33 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr13_-_29420885 | 1.32 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr7_-_30174882 | 1.31 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr4_-_5019113 | 1.24 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr3_-_16039619 | 1.22 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr6_+_25261297 | 1.19 |
ENSDART00000162824
ENSDART00000163490 ENSDART00000157790 ENSDART00000160978 ENSDART00000161545 ENSDART00000159978 |
kyat3
|
kynurenine aminotransferase 3 |
| chr6_+_39905021 | 1.17 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr7_+_38811800 | 1.17 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr9_+_28598577 | 1.14 |
ENSDART00000142623
ENSDART00000135947 |
si:ch73-7i4.1
|
si:ch73-7i4.1 |
| chr6_+_25257728 | 1.14 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr14_-_7207961 | 1.14 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr12_+_20587179 | 1.12 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr11_+_44617021 | 1.11 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr6_+_11250316 | 1.11 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr22_+_508290 | 1.11 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr7_+_23515966 | 1.10 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr17_+_24821627 | 1.10 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr23_+_16638639 | 1.09 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr17_-_25831569 | 1.08 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr12_+_36428052 | 1.06 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr16_+_25068576 | 1.06 |
ENSDART00000125838
|
im:7147486
|
im:7147486 |
| chr7_-_57509447 | 1.04 |
ENSDART00000051973
ENSDART00000147036 |
sirt3
|
sirtuin 3 |
| chr2_+_37140448 | 1.03 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr25_-_27722614 | 1.01 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr5_-_21422390 | 1.01 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr22_+_34784075 | 1.01 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr9_-_27398369 | 0.99 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr10_+_11261576 | 0.98 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr17_+_49281597 | 0.95 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr3_-_48716422 | 0.94 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr17_-_40956035 | 0.93 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr22_-_36530902 | 0.91 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr15_-_40267485 | 0.91 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr24_-_16980337 | 0.89 |
ENSDART00000183812
|
klhl15
|
kelch-like family member 15 |
| chr1_+_45663727 | 0.89 |
ENSDART00000038574
ENSDART00000141144 ENSDART00000149565 |
trappc5
|
trafficking protein particle complex 5 |
| chr2_+_2223837 | 0.89 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr1_+_51721851 | 0.88 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr5_-_11809710 | 0.87 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr9_+_54039006 | 0.85 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr4_+_17655872 | 0.84 |
ENSDART00000066999
|
washc3
|
WASH complex subunit 3 |
| chr5_-_45877387 | 0.82 |
ENSDART00000183714
ENSDART00000041503 |
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr4_-_2219705 | 0.81 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr16_+_23303859 | 0.79 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr17_+_28533102 | 0.75 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr5_-_45876908 | 0.74 |
ENSDART00000188229
|
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr7_-_72261721 | 0.71 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr21_+_27513859 | 0.70 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr9_-_32177117 | 0.70 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr13_-_38730267 | 0.68 |
ENSDART00000157524
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr24_+_23202545 | 0.67 |
ENSDART00000143862
|
pex2
|
peroxisomal biogenesis factor 2 |
| chr20_-_9095105 | 0.65 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr14_-_470505 | 0.65 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr20_+_34717403 | 0.64 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr25_-_28674739 | 0.63 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr4_+_76426308 | 0.59 |
ENSDART00000180424
ENSDART00000159789 |
FP074874.1
zgc:172128
|
zgc:172128 |
| chr1_+_47499888 | 0.57 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr12_-_31794908 | 0.57 |
ENSDART00000105583
ENSDART00000153449 |
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr17_-_2386569 | 0.55 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr20_+_23408970 | 0.53 |
ENSDART00000144261
|
fryl
|
furry homolog, like |
| chr25_-_21031007 | 0.50 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr17_+_22577472 | 0.49 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr2_+_29842878 | 0.47 |
ENSDART00000131376
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr1_-_31534089 | 0.46 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr18_-_5527050 | 0.45 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr20_+_11731039 | 0.44 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr20_-_49889111 | 0.44 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr8_-_33381430 | 0.41 |
ENSDART00000190019
|
lmx1bb
|
LIM homeobox transcription factor 1, beta b |
| chr14_-_26425416 | 0.40 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr15_-_22074315 | 0.40 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr15_+_22867174 | 0.39 |
ENSDART00000035812
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr5_-_12272689 | 0.38 |
ENSDART00000099810
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr17_-_4245902 | 0.38 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr1_+_9086942 | 0.36 |
ENSDART00000055023
|
cacng3a
|
calcium channel, voltage-dependent, gamma subunit 3a |
| chr19_+_31873308 | 0.36 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr24_-_35282568 | 0.35 |
ENSDART00000167406
ENSDART00000088609 |
sntg1
|
syntrophin, gamma 1 |
| chr24_-_7995960 | 0.35 |
ENSDART00000186594
|
bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr21_-_22635245 | 0.34 |
ENSDART00000115224
ENSDART00000101782 |
nectin1a
|
nectin cell adhesion molecule 1a |
| chr17_-_21418340 | 0.32 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr4_+_13449775 | 0.32 |
ENSDART00000172552
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr1_-_45616470 | 0.32 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr11_+_34760628 | 0.30 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr10_-_13343831 | 0.30 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr18_+_14619544 | 0.29 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr1_-_5455498 | 0.29 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr1_-_669717 | 0.28 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr21_+_38634296 | 0.28 |
ENSDART00000114467
|
AL935199.1
|
|
| chr3_+_26288981 | 0.27 |
ENSDART00000163500
|
rhot1a
|
ras homolog family member T1a |
| chr2_-_30668580 | 0.26 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr14_+_44794936 | 0.25 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr1_-_9195629 | 0.25 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr23_+_12406722 | 0.24 |
ENSDART00000145008
|
pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr25_+_13621563 | 0.22 |
ENSDART00000159428
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr10_-_11353469 | 0.21 |
ENSDART00000191847
|
lin54
|
lin-54 DREAM MuvB core complex component |
| chr4_+_19700308 | 0.20 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr25_+_22320738 | 0.17 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr1_+_52792439 | 0.16 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr1_+_11882186 | 0.16 |
ENSDART00000193419
|
snx8b
|
sorting nexin 8b |
| chr12_-_6880694 | 0.13 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr6_+_11250033 | 0.13 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr16_+_28994709 | 0.12 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr11_+_17984167 | 0.12 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr24_+_28953089 | 0.10 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr23_-_16485190 | 0.07 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr14_+_1170968 | 0.06 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr21_-_36571804 | 0.06 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr15_-_14552101 | 0.05 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr5_+_29851433 | 0.05 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr18_+_7456597 | 0.02 |
ENSDART00000142270
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr18_+_7456888 | 0.02 |
ENSDART00000081468
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr1_-_15797663 | 0.01 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr1_+_39482588 | 0.01 |
ENSDART00000181790
|
tenm3
|
teneurin transmembrane protein 3 |
| chr4_+_54618332 | 0.01 |
ENSDART00000171824
|
si:ch211-227e10.2
|
si:ch211-227e10.2 |
| chr6_-_35046735 | 0.01 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr20_+_32552912 | 0.00 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of otpa+otpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.3 | 4.0 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.7 | 2.2 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.7 | 4.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.5 | 2.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.4 | 1.3 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.4 | 2.0 | GO:0071543 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.4 | 1.5 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.3 | 3.6 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.3 | 27.0 | GO:0007599 | hemostasis(GO:0007599) |
| 0.3 | 0.8 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.3 | 1.0 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 0.7 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.2 | 0.9 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.2 | 0.9 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 2.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 1.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 2.0 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 3.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.4 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.1 | 1.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 2.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.3 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 1.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 0.7 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.7 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.4 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.8 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 1.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 5.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.4 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 9.5 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 1.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.6 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 1.4 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.5 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 1.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.8 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.8 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.0 | GO:0097240 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.6 | GO:0071696 | ectodermal placode development(GO:0071696) |
| 0.0 | 0.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 0.9 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.2 | 0.6 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 1.7 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 6.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 10.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 10.0 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.6 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.0 | 1.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 19.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 1.4 | 9.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 1.2 | 17.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.8 | 4.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.5 | 2.1 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.4 | 2.0 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.4 | 1.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 3.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 2.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.3 | 0.8 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 2.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 2.0 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 7.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.7 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 1.5 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 2.3 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 16.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.6 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 1.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.5 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 1.1 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.4 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 8.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 4.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 2.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |