Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
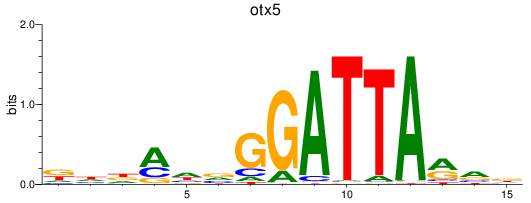
Results for otx5
Z-value: 0.69
Transcription factors associated with otx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
otx5
|
ENSDARG00000043483 | orthodenticle homolog 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx5 | dr11_v1_chr15_+_47903864_47903864 | -0.30 | 2.9e-03 | Click! |
Activity profile of otx5 motif
Sorted Z-values of otx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_3629201 | 11.37 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr15_-_5815006 | 9.21 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr14_-_41308561 | 7.35 |
ENSDART00000138232
|
arl13a
|
ADP-ribosylation factor-like 13A |
| chr4_-_17409533 | 6.00 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr4_-_8030583 | 5.90 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr20_+_53441935 | 5.85 |
ENSDART00000175214
|
apobb.2
|
apolipoprotein Bb, tandem duplicate 2 |
| chr10_+_28428222 | 5.39 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr12_-_4301234 | 4.30 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr20_-_33566640 | 4.26 |
ENSDART00000159729
|
si:dkey-65b13.9
|
si:dkey-65b13.9 |
| chr4_-_77432218 | 4.19 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr4_-_77125693 | 4.05 |
ENSDART00000174256
|
CU467646.3
|
|
| chr16_+_34111919 | 3.83 |
ENSDART00000134037
ENSDART00000006061 ENSDART00000140552 |
tcea3
|
transcription elongation factor A (SII), 3 |
| chr3_-_54544612 | 3.43 |
ENSDART00000018044
|
angptl6
|
angiopoietin-like 6 |
| chr3_+_49521106 | 3.37 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr25_+_192116 | 3.25 |
ENSDART00000153983
|
zgc:114188
|
zgc:114188 |
| chr5_-_69180587 | 3.11 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr3_+_12725343 | 3.10 |
ENSDART00000188583
|
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr11_+_29671661 | 3.08 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr4_+_18804317 | 3.08 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr16_-_26537103 | 3.03 |
ENSDART00000134908
ENSDART00000008152 |
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr4_-_77130289 | 3.03 |
ENSDART00000174380
|
CU467646.7
|
|
| chr3_+_16762483 | 3.02 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr3_+_16229911 | 2.98 |
ENSDART00000121728
|
rpl19
|
ribosomal protein L19 |
| chr23_-_31266586 | 2.85 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr10_-_1961930 | 2.63 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr11_+_18037729 | 2.62 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr21_-_20733615 | 2.58 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr23_+_44349252 | 2.55 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr24_+_10898671 | 2.46 |
ENSDART00000106272
|
si:dkey-37o8.1
|
si:dkey-37o8.1 |
| chr10_-_20445549 | 2.46 |
ENSDART00000064613
|
loxl2a
|
lysyl oxidase-like 2a |
| chr5_-_69180227 | 2.37 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr18_-_16795262 | 2.36 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr16_+_23403602 | 2.35 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr4_-_76303301 | 2.35 |
ENSDART00000158504
|
zgc:174944
|
zgc:174944 |
| chr3_+_48561112 | 2.34 |
ENSDART00000159682
|
slc16a5b
|
solute carrier family 16 (monocarboxylate transporter), member 5b |
| chr15_+_5973909 | 2.33 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr14_+_15620640 | 2.32 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr8_+_24747865 | 2.30 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr18_-_50799510 | 2.29 |
ENSDART00000174373
|
taldo1
|
transaldolase 1 |
| chr4_-_76105527 | 2.27 |
ENSDART00000159108
|
zgc:110171
|
zgc:110171 |
| chr24_-_28245872 | 2.23 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr11_-_11336986 | 2.21 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr22_-_16154771 | 2.15 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr4_-_75795899 | 2.10 |
ENSDART00000157925
|
si:dkey-261j11.1
|
si:dkey-261j11.1 |
| chr10_+_6013076 | 2.09 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr14_+_15430991 | 2.08 |
ENSDART00000158221
|
si:dkey-203a12.5
|
si:dkey-203a12.5 |
| chr4_+_20566371 | 2.07 |
ENSDART00000127576
|
BX248410.1
|
|
| chr7_-_66864756 | 2.05 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr10_-_22506044 | 2.02 |
ENSDART00000100497
|
zgc:172339
|
zgc:172339 |
| chr4_+_9508505 | 1.99 |
ENSDART00000080842
|
kitlgb
|
kit ligand b |
| chr15_+_31426768 | 1.98 |
ENSDART00000014270
|
or103-1
|
odorant receptor, family C, subfamily 103, member 1 |
| chr11_+_14287427 | 1.96 |
ENSDART00000180903
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr3_-_20957011 | 1.96 |
ENSDART00000159787
ENSDART00000180531 |
ndufa4l
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4, like |
| chr17_-_25737452 | 1.94 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr4_+_42175261 | 1.91 |
ENSDART00000162193
|
si:ch211-142b24.2
|
si:ch211-142b24.2 |
| chr2_+_7192966 | 1.91 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr23_-_45955177 | 1.90 |
ENSDART00000165963
ENSDART00000186649 ENSDART00000185773 |
LO017850.1
|
|
| chr17_+_50701748 | 1.90 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr16_-_40043322 | 1.81 |
ENSDART00000145278
|
si:dkey-29b11.3
|
si:dkey-29b11.3 |
| chr4_+_76973771 | 1.73 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr22_-_26484268 | 1.71 |
ENSDART00000110293
|
zgc:194551
|
zgc:194551 |
| chr6_-_39764995 | 1.68 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_-_26049282 | 1.68 |
ENSDART00000136389
ENSDART00000101124 |
rnaseka
|
ribonuclease, RNase K a |
| chr9_+_18829360 | 1.65 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr22_+_25140904 | 1.62 |
ENSDART00000143114
|
si:dkeyp-20e4.8
|
si:dkeyp-20e4.8 |
| chr10_-_36738619 | 1.60 |
ENSDART00000093000
ENSDART00000157179 |
PLEKHB1
|
si:ch211-176g13.7 |
| chr22_-_8509215 | 1.60 |
ENSDART00000140146
|
si:ch73-27e22.3
|
si:ch73-27e22.3 |
| chr9_+_1313418 | 1.57 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr15_-_36727462 | 1.56 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr16_-_25741225 | 1.56 |
ENSDART00000130641
|
bcl3
|
B cell CLL/lymphoma 3 |
| chr18_-_40684756 | 1.55 |
ENSDART00000113799
ENSDART00000139042 |
si:ch211-132b12.7
|
si:ch211-132b12.7 |
| chr25_+_19041329 | 1.55 |
ENSDART00000153467
|
lrtm2b
|
leucine-rich repeats and transmembrane domains 2b |
| chr19_-_2115040 | 1.55 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr4_+_25558849 | 1.51 |
ENSDART00000113663
ENSDART00000100755 ENSDART00000111416 ENSDART00000127840 ENSDART00000168618 ENSDART00000111820 ENSDART00000113866 ENSDART00000110107 ENSDART00000111344 ENSDART00000108548 |
zgc:195175
|
zgc:195175 |
| chr8_+_6954984 | 1.51 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr20_+_51478939 | 1.51 |
ENSDART00000149758
|
tlr5a
|
toll-like receptor 5a |
| chr4_+_62431132 | 1.50 |
ENSDART00000160226
|
si:ch211-79g12.1
|
si:ch211-79g12.1 |
| chr23_+_39854566 | 1.50 |
ENSDART00000190423
ENSDART00000164473 ENSDART00000161881 |
si:ch73-217b7.1
|
si:ch73-217b7.1 |
| chr20_+_9128829 | 1.50 |
ENSDART00000064144
ENSDART00000137450 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr25_-_27819838 | 1.49 |
ENSDART00000067106
|
lmod2a
|
leiomodin 2 (cardiac) a |
| chr21_-_43428040 | 1.49 |
ENSDART00000148325
|
stk26
|
serine/threonine protein kinase 26 |
| chr4_-_13614797 | 1.48 |
ENSDART00000138366
ENSDART00000165212 |
irf5
|
interferon regulatory factor 5 |
| chr19_-_3261019 | 1.46 |
ENSDART00000134407
|
si:ch211-133n4.12
|
si:ch211-133n4.12 |
| chr23_+_39963599 | 1.46 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr9_-_12574473 | 1.46 |
ENSDART00000191372
ENSDART00000193667 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr18_+_48758916 | 1.45 |
ENSDART00000060098
|
gemin7
|
gem (nuclear organelle) associated protein 7 |
| chr18_+_33490315 | 1.45 |
ENSDART00000140885
ENSDART00000155210 |
v2rh1
|
vomeronasal 2 receptor, h1 |
| chr3_+_46764278 | 1.43 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr19_+_43715911 | 1.42 |
ENSDART00000006344
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr25_-_17552299 | 1.40 |
ENSDART00000154327
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr1_+_8110562 | 1.38 |
ENSDART00000112160
|
SLC5A10
|
si:dkeyp-9d4.5 |
| chr14_+_2487672 | 1.38 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr18_-_42071876 | 1.37 |
ENSDART00000193288
ENSDART00000189021 ENSDART00000019245 |
arhgap42b
|
Rho GTPase activating protein 42b |
| chr14_+_31751260 | 1.37 |
ENSDART00000169796
|
si:dkeyp-11e3.1
|
si:dkeyp-11e3.1 |
| chr11_-_3366782 | 1.35 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr16_+_23404170 | 1.34 |
ENSDART00000170061
|
s100w
|
S100 calcium binding protein W |
| chr17_+_4427463 | 1.34 |
ENSDART00000131593
|
si:zfos-364h11.2
|
si:zfos-364h11.2 |
| chr16_+_44906324 | 1.32 |
ENSDART00000074960
|
cd22
|
cd22 molecule |
| chr4_+_58667348 | 1.32 |
ENSDART00000186596
ENSDART00000180673 |
CR388148.2
|
|
| chr9_+_20519846 | 1.31 |
ENSDART00000109680
ENSDART00000142220 |
vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr4_-_14470071 | 1.31 |
ENSDART00000143773
|
plxnb2a
|
plexin b2a |
| chr3_-_54524194 | 1.30 |
ENSDART00000155406
ENSDART00000111791 |
si:ch73-208g10.1
|
si:ch73-208g10.1 |
| chr25_+_469855 | 1.29 |
ENSDART00000104717
|
rsl24d1
|
ribosomal L24 domain containing 1 |
| chr3_-_27647845 | 1.24 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr11_+_6136220 | 1.23 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr16_+_52343905 | 1.20 |
ENSDART00000131051
|
ifnlr1
|
interferon lambda receptor 1 |
| chr2_-_31686353 | 1.20 |
ENSDART00000126177
ENSDART00000056679 |
e2f5
|
E2F transcription factor 5 |
| chr17_-_8592824 | 1.19 |
ENSDART00000127022
|
CU462878.1
|
|
| chr4_+_18441988 | 1.15 |
ENSDART00000040827
|
ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr4_+_36616118 | 1.14 |
ENSDART00000160058
ENSDART00000170506 |
si:dkey-151g22.1
|
si:dkey-151g22.1 |
| chr4_-_71554068 | 1.13 |
ENSDART00000167414
|
si:dkey-27n6.1
|
si:dkey-27n6.1 |
| chr23_+_7505943 | 1.13 |
ENSDART00000135787
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr10_-_1961576 | 1.12 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr4_+_43245188 | 1.11 |
ENSDART00000150310
|
si:dkey-29j8.1
|
si:dkey-29j8.1 |
| chr8_-_2543327 | 1.11 |
ENSDART00000134115
|
si:ch211-51h9.6
|
si:ch211-51h9.6 |
| chr24_+_17005647 | 1.09 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr11_-_15090564 | 1.09 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr18_+_27218737 | 1.09 |
ENSDART00000111450
|
plekha7a
|
pleckstrin homology domain containing, family A member 7a |
| chr4_-_49133107 | 1.08 |
ENSDART00000150806
|
znf1146
|
zinc finger protein 1146 |
| chr14_+_15191176 | 1.07 |
ENSDART00000183447
ENSDART00000193093 ENSDART00000169309 |
si:dkey-203a12.2
|
si:dkey-203a12.2 |
| chr3_-_25119839 | 1.06 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr4_+_75575252 | 1.06 |
ENSDART00000166536
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr4_-_76186742 | 1.05 |
ENSDART00000163970
|
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr20_+_6533260 | 1.05 |
ENSDART00000135005
ENSDART00000166356 |
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr11_+_31558207 | 1.05 |
ENSDART00000140204
|
egln1b
|
egl-9 family hypoxia-inducible factor 1b |
| chr4_+_70341246 | 1.04 |
ENSDART00000123408
|
si:ch211-76m11.3
|
si:ch211-76m11.3 |
| chr19_-_9760994 | 1.04 |
ENSDART00000139727
|
si:dkey-14o18.2
|
si:dkey-14o18.2 |
| chr23_+_25291891 | 1.00 |
ENSDART00000016248
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr8_+_2656231 | 1.00 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr8_+_21297217 | 1.00 |
ENSDART00000142836
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr14_-_1280907 | 1.00 |
ENSDART00000186150
|
CABZ01081490.1
|
|
| chr2_+_51783120 | 0.99 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr12_+_9320783 | 0.97 |
ENSDART00000152526
|
kcnh6b
|
potassium voltage-gated channel, subfamily H (eag-related), member 6b |
| chr1_+_8111009 | 0.96 |
ENSDART00000152192
|
SLC5A10
|
si:dkeyp-9d4.5 |
| chr9_+_3519191 | 0.96 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr4_-_49884959 | 0.95 |
ENSDART00000190248
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr1_-_46859398 | 0.95 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr12_-_30777540 | 0.94 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr6_+_46481024 | 0.93 |
ENSDART00000155596
|
si:dkey-7k24.5
|
si:dkey-7k24.5 |
| chr4_-_71471302 | 0.91 |
ENSDART00000187687
|
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr24_+_36317544 | 0.91 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr11_+_31680513 | 0.90 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr20_-_45722895 | 0.90 |
ENSDART00000153273
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr17_+_16046132 | 0.89 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr1_+_29515160 | 0.89 |
ENSDART00000152226
|
si:dkey-228l14.1
|
si:dkey-228l14.1 |
| chr4_+_43225728 | 0.89 |
ENSDART00000158639
ENSDART00000141339 ENSDART00000137770 ENSDART00000166019 |
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr22_+_38164486 | 0.88 |
ENSDART00000137521
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr20_+_30725778 | 0.88 |
ENSDART00000062532
|
nhsl1b
|
NHS-like 1b |
| chr22_-_26005894 | 0.87 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr24_+_19518303 | 0.87 |
ENSDART00000027022
ENSDART00000056080 |
sulf1
|
sulfatase 1 |
| chr4_-_71214516 | 0.86 |
ENSDART00000169997
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr13_+_25486608 | 0.85 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr10_+_19525839 | 0.85 |
ENSDART00000162912
ENSDART00000158512 |
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr18_+_33353755 | 0.85 |
ENSDART00000144160
|
v2rh13
|
vomeronasal 2 receptor, h13 |
| chr6_-_39521832 | 0.85 |
ENSDART00000065038
|
atf1
|
activating transcription factor 1 |
| chr18_+_45862414 | 0.84 |
ENSDART00000024615
|
rnpepl1
|
arginyl aminopeptidase like 1 |
| chr15_-_29510074 | 0.83 |
ENSDART00000184070
|
BX324227.3
|
|
| chr9_+_343062 | 0.83 |
ENSDART00000164012
ENSDART00000164365 |
bpifcl
|
BPI fold containing family C, like |
| chr7_+_8456999 | 0.82 |
ENSDART00000172880
|
jac4
|
jacalin 4 |
| chr4_-_52783184 | 0.82 |
ENSDART00000172283
|
si:dkey-4j21.2
|
si:dkey-4j21.2 |
| chr17_+_31237866 | 0.80 |
ENSDART00000076943
|
prlh2r
|
prolactin releasing hormone 2 receptor |
| chr4_-_39186893 | 0.79 |
ENSDART00000150371
ENSDART00000121804 ENSDART00000133901 |
si:ch211-22k7.9
|
si:ch211-22k7.9 |
| chr6_+_32834760 | 0.79 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr22_-_12726556 | 0.79 |
ENSDART00000105776
|
fam207a
|
family with sequence similarity 207, member A |
| chr1_-_58009216 | 0.77 |
ENSDART00000143829
|
nxnl1
|
nucleoredoxin like 1 |
| chr3_+_14157090 | 0.76 |
ENSDART00000156766
|
si:ch211-108d22.2
|
si:ch211-108d22.2 |
| chr5_-_1827575 | 0.76 |
ENSDART00000171193
|
mcc
|
mutated in colorectal cancers |
| chr4_-_33763221 | 0.76 |
ENSDART00000132613
ENSDART00000191816 ENSDART00000188473 |
si:ch211-197f20.1
|
si:ch211-197f20.1 |
| chr4_-_56103062 | 0.75 |
ENSDART00000165784
ENSDART00000190846 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr3_+_23703704 | 0.74 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr18_+_2189211 | 0.74 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr4_+_58661331 | 0.74 |
ENSDART00000188763
|
CR388148.2
|
|
| chr3_+_26144765 | 0.74 |
ENSDART00000146267
ENSDART00000043932 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr12_-_16558106 | 0.74 |
ENSDART00000109033
|
si:dkey-269i1.4
|
si:dkey-269i1.4 |
| chr4_-_61419027 | 0.73 |
ENSDART00000181567
ENSDART00000187397 ENSDART00000180671 ENSDART00000164053 ENSDART00000187395 |
znf1021
|
zinc finger protein 1021 |
| chr4_+_51201875 | 0.72 |
ENSDART00000189875
ENSDART00000186722 |
BX005380.1
|
|
| chr22_-_26100282 | 0.72 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr4_+_67994238 | 0.71 |
ENSDART00000160968
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
| chr4_-_34459615 | 0.71 |
ENSDART00000169144
ENSDART00000167022 |
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr4_-_43919359 | 0.71 |
ENSDART00000184163
ENSDART00000158019 ENSDART00000191745 ENSDART00000181638 ENSDART00000150827 |
znf1104
|
zinc finger protein 1104 |
| chr22_-_5006119 | 0.69 |
ENSDART00000187998
|
rx1
|
retinal homeobox gene 1 |
| chr12_+_31537604 | 0.68 |
ENSDART00000153340
|
si:ch73-205h11.1
|
si:ch73-205h11.1 |
| chr13_+_22863516 | 0.67 |
ENSDART00000113082
ENSDART00000189200 |
hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr9_-_3519253 | 0.67 |
ENSDART00000169586
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr4_-_34460301 | 0.67 |
ENSDART00000192977
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr21_+_5080789 | 0.67 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr3_+_24094581 | 0.67 |
ENSDART00000138270
ENSDART00000131509 |
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr20_+_29587995 | 0.67 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr24_+_14527935 | 0.66 |
ENSDART00000134846
|
si:dkeyp-73g8.5
|
si:dkeyp-73g8.5 |
| chr4_-_47095583 | 0.66 |
ENSDART00000183595
ENSDART00000183279 ENSDART00000191638 |
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr3_-_26183699 | 0.65 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr5_-_30074332 | 0.65 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr4_+_57192681 | 0.65 |
ENSDART00000190438
|
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr4_-_67984478 | 0.65 |
ENSDART00000160623
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr4_+_46812891 | 0.65 |
ENSDART00000189655
ENSDART00000162329 |
si:ch211-215p11.3
|
si:ch211-215p11.3 |
| chr4_+_35227135 | 0.65 |
ENSDART00000161023
ENSDART00000182403 |
si:ch211-276i6.1
|
si:ch211-276i6.1 |
| chr4_+_60547709 | 0.65 |
ENSDART00000158732
|
si:dkey-211i20.5
|
si:dkey-211i20.5 |
| chr4_-_63559311 | 0.64 |
ENSDART00000164415
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr12_-_30777743 | 0.64 |
ENSDART00000148888
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr7_-_17779644 | 0.64 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of otx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 1.2 | 3.7 | GO:0090008 | hypoblast development(GO:0090008) |
| 1.2 | 5.8 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.8 | 2.4 | GO:0010656 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.7 | 2.1 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.6 | 2.5 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.5 | 6.0 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.5 | 2.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.4 | 1.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 1.2 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.4 | 1.6 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.4 | 1.5 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.4 | 1.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 1.6 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.3 | 1.5 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 1.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 1.6 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.2 | 4.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.2 | 2.3 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.2 | 4.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.7 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 2.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 2.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 2.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 3.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 3.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.1 | 3.0 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 0.5 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.5 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 0.6 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 1.4 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 0.9 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.7 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.6 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 6.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.3 | GO:0015884 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.2 | GO:0002828 | regulation of type 2 immune response(GO:0002828) |
| 0.1 | 0.7 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 2.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.5 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 1.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 3.1 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.9 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 1.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 4.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.6 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.9 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.6 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 1.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.9 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 4.2 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.4 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.5 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.0 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 1.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 2.3 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.3 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.1 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 1.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 1.7 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.3 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 1.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 5.9 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 1.7 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 29.5 | GO:0035556 | intracellular signal transduction(GO:0035556) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.8 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.8 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.4 | 1.4 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.3 | 1.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 5.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.2 | 1.6 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.2 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 1.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.2 | 1.4 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 2.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 1.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.7 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 4.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.8 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 4.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 5.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 2.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.7 | 2.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.7 | 6.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.6 | 5.8 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.5 | 4.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.5 | 2.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.5 | 1.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 4.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 2.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.4 | 3.0 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.4 | 1.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 0.9 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.3 | 3.7 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.2 | 1.0 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 0.6 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.2 | 3.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 0.6 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.2 | 1.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 0.5 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 1.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 0.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 3.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.6 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 2.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 6.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 3.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.4 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 0.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.3 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.1 | 5.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 2.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 2.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.7 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.5 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.1 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 3.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 7.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.2 | 4.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 1.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 1.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 3.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 1.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 1.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 2.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 6.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 1.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |