Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
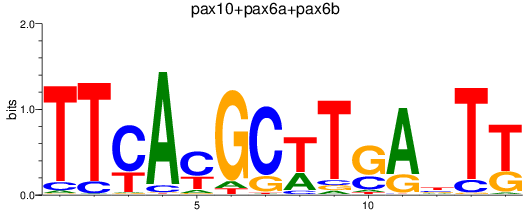
Results for pax10+pax6a+pax6b
Z-value: 0.78
Transcription factors associated with pax10+pax6a+pax6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax6b
|
ENSDARG00000045936 | paired box 6b |
|
pax10
|
ENSDARG00000053364 | paired box 10 |
|
pax6a
|
ENSDARG00000103379 | paired box 6a |
|
pax10
|
ENSDARG00000111379 | paired box 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax10 | dr11_v1_chr3_+_32553714_32553714 | -0.37 | 2.9e-04 | Click! |
| pax6b | dr11_v1_chr7_+_15872357_15872357 | -0.33 | 1.3e-03 | Click! |
| pax6a | dr11_v1_chr25_-_15049694_15049781 | -0.32 | 1.9e-03 | Click! |
Activity profile of pax10+pax6a+pax6b motif
Sorted Z-values of pax10+pax6a+pax6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_38306010 | 12.15 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr10_-_8053385 | 10.41 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr10_-_8046764 | 9.81 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr10_-_8053753 | 7.00 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr7_+_38750871 | 6.45 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr13_-_22862133 | 6.08 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr21_+_27382893 | 5.92 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr2_+_6255434 | 5.73 |
ENSDART00000139429
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr25_+_22320738 | 5.68 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr23_+_17926279 | 5.05 |
ENSDART00000012540
|
chia.4
|
chitinase, acidic.4 |
| chr9_-_32847642 | 4.49 |
ENSDART00000121506
|
hpx
|
hemopexin |
| chr5_+_57924611 | 4.46 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr7_-_20582842 | 4.26 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr23_+_44611864 | 4.21 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr9_+_8365398 | 4.03 |
ENSDART00000138713
ENSDART00000136847 |
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr3_+_43086548 | 3.73 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr6_+_23887314 | 3.58 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr14_-_1454045 | 3.26 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr13_-_20381485 | 3.06 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr8_+_52442622 | 3.00 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr9_-_1970071 | 2.92 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr15_-_12270857 | 2.92 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr25_-_31423493 | 2.90 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr1_+_5402476 | 2.75 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr10_+_25726694 | 2.74 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr13_+_10945337 | 2.55 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr15_+_1534644 | 2.53 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr23_+_44614056 | 2.52 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr9_+_23003208 | 2.52 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr21_-_2322102 | 2.51 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr20_-_25486384 | 2.48 |
ENSDART00000141340
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr7_+_3357973 | 2.39 |
ENSDART00000172853
|
si:ch211-285c6.5
|
si:ch211-285c6.5 |
| chr25_-_27842654 | 2.34 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr18_+_62932 | 2.25 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr14_-_858985 | 2.25 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr18_+_14277003 | 2.25 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr19_+_30990815 | 2.23 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr19_-_4793263 | 2.19 |
ENSDART00000147510
ENSDART00000141336 ENSDART00000110551 ENSDART00000146684 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr21_-_25756119 | 2.19 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr12_+_25845494 | 2.13 |
ENSDART00000170582
|
glud1b
|
glutamate dehydrogenase 1b |
| chr10_-_17501528 | 2.09 |
ENSDART00000144847
|
slc2a11l
|
solute carrier family 2 (facilitated glucose transporter), member 11-like |
| chr13_-_10945288 | 2.07 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr14_-_14659023 | 2.01 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr9_-_42873700 | 2.00 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr15_-_1745408 | 1.99 |
ENSDART00000182311
|
stx1a
|
syntaxin 1A (brain) |
| chr19_-_11238620 | 1.96 |
ENSDART00000148697
ENSDART00000149169 |
ssr2
|
signal sequence receptor, beta |
| chr16_+_13993285 | 1.94 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr13_-_25196758 | 1.91 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr2_+_21000334 | 1.90 |
ENSDART00000062563
ENSDART00000147809 |
rreb1b
|
ras responsive element binding protein 1b |
| chr2_-_21349425 | 1.89 |
ENSDART00000171699
|
hhatla
|
hedgehog acyltransferase like, a |
| chr13_+_12671513 | 1.88 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr17_+_8799661 | 1.86 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr4_-_18850799 | 1.86 |
ENSDART00000151844
|
mcat
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr20_+_1996202 | 1.80 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr1_+_51721851 | 1.77 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr1_+_12068800 | 1.74 |
ENSDART00000055566
|
xpa
|
xeroderma pigmentosum, complementation group A |
| chr8_+_52415603 | 1.71 |
ENSDART00000021604
ENSDART00000191424 |
gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr25_-_37284370 | 1.71 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr8_+_41038141 | 1.69 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr25_-_4148719 | 1.68 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr2_+_21048661 | 1.65 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr2_+_11031360 | 1.62 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr24_-_36727922 | 1.62 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr18_+_45781516 | 1.62 |
ENSDART00000168840
|
rpl35a
|
ribosomal protein L35a |
| chr12_-_48671612 | 1.60 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr11_+_25619892 | 1.59 |
ENSDART00000146647
ENSDART00000033702 |
comtb
|
catechol-O-methyltransferase b |
| chr16_+_11779534 | 1.59 |
ENSDART00000133497
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr18_+_45781115 | 1.58 |
ENSDART00000151699
ENSDART00000179887 |
rpl35a
|
ribosomal protein L35a |
| chr11_-_3897067 | 1.50 |
ENSDART00000134858
|
rpn1
|
ribophorin I |
| chr16_-_13992646 | 1.45 |
ENSDART00000139623
|
si:dkey-85k15.6
|
si:dkey-85k15.6 |
| chr7_+_59649746 | 1.45 |
ENSDART00000181067
|
rpl34
|
ribosomal protein L34 |
| chr21_-_39566854 | 1.40 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr15_-_14625373 | 1.35 |
ENSDART00000171841
|
slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr10_-_25823258 | 1.34 |
ENSDART00000064327
|
ftr54
|
finTRIM family, member 54 |
| chr9_+_40546677 | 1.34 |
ENSDART00000132911
|
vwc2l
|
von Willebrand factor C domain containing protein 2-like |
| chr25_+_37285737 | 1.31 |
ENSDART00000126879
|
tmed6
|
transmembrane p24 trafficking protein 6 |
| chr15_-_44663712 | 1.31 |
ENSDART00000192124
|
CABZ01061894.1
|
|
| chr7_-_45852270 | 1.30 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr13_-_39160018 | 1.30 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr10_-_31015535 | 1.29 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr8_+_41037541 | 1.29 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr13_-_39159810 | 1.25 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr18_-_24988645 | 1.22 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr23_+_36052944 | 1.20 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| chr4_-_17741513 | 1.18 |
ENSDART00000141680
|
mybpc1
|
myosin binding protein C, slow type |
| chr12_+_47794089 | 1.15 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr11_-_11336986 | 1.15 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr22_+_8365905 | 1.13 |
ENSDART00000105945
|
si:dkey-19a16.11
|
si:dkey-19a16.11 |
| chr24_+_26379441 | 1.12 |
ENSDART00000137786
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr9_+_31222026 | 1.10 |
ENSDART00000145573
|
clybl
|
citrate lyase beta like |
| chr11_-_45152912 | 1.09 |
ENSDART00000167540
ENSDART00000173050 ENSDART00000170795 |
afmid
|
arylformamidase |
| chr16_+_11779761 | 1.08 |
ENSDART00000140297
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr24_+_28561038 | 1.08 |
ENSDART00000147063
|
abca4a
|
ATP-binding cassette, sub-family A (ABC1), member 4a |
| chr21_+_23108420 | 1.08 |
ENSDART00000192394
ENSDART00000088459 |
htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr5_-_4204580 | 1.07 |
ENSDART00000049197
ENSDART00000132130 |
si:ch211-283g2.1
|
si:ch211-283g2.1 |
| chr1_+_9199031 | 1.07 |
ENSDART00000092058
ENSDART00000182771 |
chtf18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
| chr24_-_2381143 | 1.06 |
ENSDART00000144307
|
rreb1a
|
ras responsive element binding protein 1a |
| chr9_-_50000144 | 1.05 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr18_-_14901437 | 1.02 |
ENSDART00000145842
ENSDART00000008035 |
trabd
|
TraB domain containing |
| chr10_+_26972755 | 1.02 |
ENSDART00000042162
|
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr2_+_28453338 | 1.02 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr20_+_53474963 | 1.01 |
ENSDART00000138976
|
bub1ba
|
BUB1 mitotic checkpoint serine/threonine kinase Ba |
| chr8_+_37111643 | 1.01 |
ENSDART00000061336
|
ren
|
renin |
| chr17_+_8799451 | 1.00 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr1_+_11107688 | 0.99 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr18_-_17513426 | 0.99 |
ENSDART00000146725
|
b3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr1_-_54706039 | 0.98 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr7_+_59649399 | 0.98 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr3_-_23513155 | 0.97 |
ENSDART00000170200
|
BX682558.1
|
|
| chr22_-_5006801 | 0.97 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr12_+_4900981 | 0.94 |
ENSDART00000171507
|
cd79b
|
CD79b molecule, immunoglobulin-associated beta |
| chr11_+_6115621 | 0.94 |
ENSDART00000165031
ENSDART00000027666 ENSDART00000161458 |
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr23_-_9925568 | 0.93 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr19_-_25114701 | 0.91 |
ENSDART00000149035
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr3_-_54607166 | 0.90 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr8_-_8489886 | 0.89 |
ENSDART00000183334
|
abt1
|
activator of basal transcription 1 |
| chr23_-_27235403 | 0.89 |
ENSDART00000134418
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr4_-_5370468 | 0.89 |
ENSDART00000133423
ENSDART00000184963 |
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr21_+_43402794 | 0.89 |
ENSDART00000185572
|
frmd7
|
FERM domain containing 7 |
| chr22_-_3182965 | 0.88 |
ENSDART00000158009
|
lonp1
|
lon peptidase 1, mitochondrial |
| chr24_+_39186940 | 0.87 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr22_+_9091913 | 0.86 |
ENSDART00000186598
|
BX248395.1
|
|
| chr14_-_30918662 | 0.86 |
ENSDART00000176631
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr11_-_45152702 | 0.85 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr8_+_52442785 | 0.85 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr9_+_23900703 | 0.84 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr22_+_22438783 | 0.84 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr9_-_19583704 | 0.84 |
ENSDART00000053218
|
tmprss3a
|
transmembrane protease, serine 3a |
| chr17_-_12196865 | 0.84 |
ENSDART00000154694
|
kif28
|
kinesin family member 28 |
| chr5_-_25072607 | 0.83 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr21_-_1644414 | 0.83 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr7_-_58244220 | 0.82 |
ENSDART00000180450
|
unm_hu7910
|
un-named hu7910 |
| chr20_+_37393134 | 0.82 |
ENSDART00000128321
|
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr22_+_21398508 | 0.82 |
ENSDART00000089408
ENSDART00000186091 |
shdb
|
Src homology 2 domain containing transforming protein D, b |
| chr2_+_6991208 | 0.82 |
ENSDART00000182239
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr20_-_3238110 | 0.81 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr19_-_20163197 | 0.79 |
ENSDART00000148179
ENSDART00000151646 |
fam221a
|
family with sequence similarity 221, member A |
| chr3_-_8542577 | 0.78 |
ENSDART00000183527
|
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr19_-_15192840 | 0.78 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr14_-_12390724 | 0.78 |
ENSDART00000131343
|
magt1
|
magnesium transporter 1 |
| chr2_-_32768951 | 0.77 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr20_+_19119800 | 0.77 |
ENSDART00000151881
|
rp1l1b
|
retinitis pigmentosa 1-like 1b |
| chr3_-_26184018 | 0.77 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr6_-_58757131 | 0.77 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr11_-_165288 | 0.77 |
ENSDART00000108703
ENSDART00000173151 |
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
| chr13_-_11378127 | 0.77 |
ENSDART00000158632
|
akt3a
|
v-akt murine thymoma viral oncogene homolog 3a |
| chr7_+_26545502 | 0.77 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr5_-_55914268 | 0.76 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr7_+_27250186 | 0.75 |
ENSDART00000150068
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr21_-_3700334 | 0.75 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr9_-_1949915 | 0.75 |
ENSDART00000190712
|
hoxd3a
|
homeobox D3a |
| chr23_+_38957738 | 0.75 |
ENSDART00000193480
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr24_-_26479841 | 0.75 |
ENSDART00000079984
|
rpl22l1
|
ribosomal protein L22-like 1 |
| chr18_-_20466061 | 0.75 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr4_+_18804317 | 0.74 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr25_+_23280220 | 0.74 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr4_-_13613148 | 0.74 |
ENSDART00000067164
ENSDART00000111247 |
irf5
|
interferon regulatory factor 5 |
| chr20_-_47348116 | 0.74 |
ENSDART00000162087
ENSDART00000160769 ENSDART00000164484 |
dtnba
|
dystrobrevin, beta a |
| chr8_+_38417461 | 0.73 |
ENSDART00000132718
|
nkx6.3
|
NK6 homeobox 3 |
| chr13_-_50200042 | 0.73 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr10_-_19497914 | 0.72 |
ENSDART00000132084
|
si:ch211-127i16.2
|
si:ch211-127i16.2 |
| chr25_-_28926631 | 0.72 |
ENSDART00000112850
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr9_-_746317 | 0.72 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr9_-_2936017 | 0.72 |
ENSDART00000102823
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr10_-_45002196 | 0.71 |
ENSDART00000170767
|
ved
|
ventrally expressed dharma/bozozok antagonist |
| chr24_-_1010467 | 0.71 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr12_-_19250854 | 0.70 |
ENSDART00000152844
|
cdc42ep1a
|
CDC42 effector protein (Rho GTPase binding) 1a |
| chr17_-_27266053 | 0.68 |
ENSDART00000110903
|
E2F2
|
si:ch211-160f23.5 |
| chr1_+_55239160 | 0.67 |
ENSDART00000152318
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr6_-_39344259 | 0.67 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr11_+_12897004 | 0.67 |
ENSDART00000152007
|
si:dkey-11m19.5
|
si:dkey-11m19.5 |
| chr12_-_16595406 | 0.66 |
ENSDART00000166798
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr17_-_36841981 | 0.66 |
ENSDART00000131566
|
myo6b
|
myosin VIb |
| chr7_-_1101071 | 0.66 |
ENSDART00000176053
|
dctn1a
|
dynactin 1a |
| chr25_-_7759453 | 0.65 |
ENSDART00000142439
ENSDART00000021577 |
phf21ab
|
PHD finger protein 21Ab |
| chr8_-_45729248 | 0.65 |
ENSDART00000187736
|
BX088696.1
|
|
| chr23_+_38957472 | 0.65 |
ENSDART00000193836
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr25_+_30105628 | 0.65 |
ENSDART00000187704
|
si:ch211-147k10.5
|
si:ch211-147k10.5 |
| chr25_-_10543682 | 0.65 |
ENSDART00000154262
|
tesmin
|
testis expressed metallothionein like protein |
| chr10_+_6625785 | 0.65 |
ENSDART00000148433
|
si:ch211-57m13.3
|
si:ch211-57m13.3 |
| chr14_+_36454087 | 0.64 |
ENSDART00000180247
|
TENM3
|
si:dkey-237h12.3 |
| chr5_-_26247973 | 0.64 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr24_+_37709191 | 0.64 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr10_-_167782 | 0.62 |
ENSDART00000108780
|
erg
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr3_-_25118777 | 0.62 |
ENSDART00000188222
|
chadla
|
chondroadherin-like a |
| chr7_-_3850090 | 0.62 |
ENSDART00000113403
ENSDART00000137683 ENSDART00000049926 |
si:dkey-28d5.12
si:dkey-88n24.7
|
si:dkey-28d5.12 si:dkey-88n24.7 |
| chr17_-_50018133 | 0.62 |
ENSDART00000112267
|
filip1a
|
filamin A interacting protein 1a |
| chr11_-_8262105 | 0.61 |
ENSDART00000173225
|
si:cabz01021066.1
|
si:cabz01021066.1 |
| chr14_+_7902374 | 0.61 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr23_+_25292147 | 0.61 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr1_+_33322174 | 0.61 |
ENSDART00000140435
|
mxra5a
|
matrix-remodelling associated 5a |
| chr9_+_7732714 | 0.61 |
ENSDART00000145853
|
si:ch1073-349o24.2
|
si:ch1073-349o24.2 |
| chr2_+_17569644 | 0.61 |
ENSDART00000135823
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr4_+_56477160 | 0.60 |
ENSDART00000184152
|
si:ch211-237a4.2
|
si:ch211-237a4.2 |
| chr16_-_5105295 | 0.60 |
ENSDART00000082071
ENSDART00000148955 ENSDART00000184700 ENSDART00000188127 |
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr8_-_45729758 | 0.59 |
ENSDART00000189329
|
BX088696.1
|
|
| chr3_-_34717882 | 0.59 |
ENSDART00000151127
|
thraa
|
thyroid hormone receptor alpha a |
| chr12_-_35582683 | 0.59 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr10_-_22497980 | 0.58 |
ENSDART00000185549
|
CT573337.2
|
|
| chr2_+_38731696 | 0.58 |
ENSDART00000181733
|
FQ377660.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of pax10+pax6a+pax6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.7 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 1.0 | 2.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.9 | 6.5 | GO:1900048 | acute-phase response(GO:0006953) positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.8 | 6.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.6 | 1.9 | GO:1903060 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.6 | 1.7 | GO:0070914 | UV-damage excision repair(GO:0070914) nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.5 | 1.5 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.5 | 5.1 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.4 | 3.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.4 | 2.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.3 | 5.7 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 1.9 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.3 | 1.9 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 2.1 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.3 | 5.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 1.7 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.3 | 0.8 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.2 | 1.9 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.2 | 0.7 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.2 | 1.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 1.6 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.2 | 1.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 2.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 0.8 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.2 | 1.3 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.2 | 0.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 1.8 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 3.9 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.2 | 0.8 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 1.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 4.6 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 2.9 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 2.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.5 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.4 | GO:0006168 | adenine salvage(GO:0006168) guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.1 | 2.3 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 2.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.6 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 2.5 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 0.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.3 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.2 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.1 | 0.7 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.6 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) regulation of fatty acid oxidation(GO:0046320) |
| 0.1 | 0.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.8 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.2 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.1 | 0.4 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 1.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.7 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 1.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.7 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.6 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.6 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.9 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 2.0 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.5 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 0.7 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 1.0 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 2.6 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 2.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 3.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 4.3 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.8 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 11.5 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.0 | 0.7 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 3.7 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.8 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.0 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 1.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.8 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.6 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 1.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 1.9 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 1.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.4 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.7 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.7 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.8 | GO:0042461 | photoreceptor cell development(GO:0042461) |
| 0.0 | 0.1 | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902253) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.7 | 27.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 1.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.4 | 2.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 1.7 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 2.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 0.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 3.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.6 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 0.5 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.2 | 2.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.2 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.6 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 1.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.8 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.0 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 2.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 3.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 2.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 6.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.8 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 4.2 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.9 | 27.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.8 | 6.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 1.9 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.6 | 1.9 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.5 | 5.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.5 | 5.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.4 | 2.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.4 | 2.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.4 | 1.6 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.4 | 1.9 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.4 | 2.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.4 | 3.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.4 | 1.1 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.3 | 3.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 2.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 1.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 1.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 2.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 0.8 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 0.8 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 2.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 0.7 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 2.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.4 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 0.5 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 1.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.5 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.1 | GO:0043142 | DNA clamp loader activity(GO:0003689) single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) protein-DNA loading ATPase activity(GO:0033170) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 1.7 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 0.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 4.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 2.1 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 0.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 1.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 2.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.0 | 1.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 7.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 6.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 4.3 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 0.6 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 4.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 3.4 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 3.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 5.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 2.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.7 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 2.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.7 | 2.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.6 | 5.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 4.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 6.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 5.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 1.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.6 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 1.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 9.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |