Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
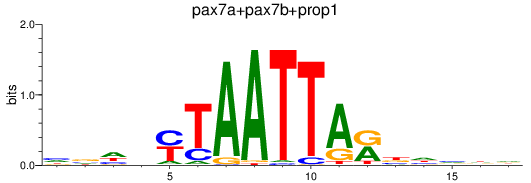
Results for pax7a+pax7b+prop1
Z-value: 0.59
Transcription factors associated with pax7a+pax7b+prop1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prop1
|
ENSDARG00000039756 | PROP paired-like homeobox 1 |
|
pax7b
|
ENSDARG00000070818 | paired box 7b |
|
pax7a
|
ENSDARG00000100398 | paired box 7a |
|
prop1
|
ENSDARG00000109950 | PROP paired-like homeobox 1 |
|
pax7a
|
ENSDARG00000111233 | paired box 7a |
|
prop1
|
ENSDARG00000114449 | PROP paired-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax7b | dr11_v1_chr23_+_20863145_20863145 | 0.21 | 3.8e-02 | Click! |
| pax7a | dr11_v1_chr11_+_41242644_41242644 | 0.21 | 3.8e-02 | Click! |
| prop1 | dr11_v1_chr13_+_8677166_8677304 | -0.01 | 9.5e-01 | Click! |
Activity profile of pax7a+pax7b+prop1 motif
Sorted Z-values of pax7a+pax7b+prop1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_1012269 | 4.36 |
ENSDART00000159835
|
CABZ01057159.1
|
|
| chr6_-_58764672 | 4.32 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr5_+_2815021 | 3.99 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr2_+_6253246 | 3.32 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr20_-_40755614 | 3.31 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr10_-_34002185 | 3.14 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr9_+_8396755 | 3.08 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr20_+_54304800 | 2.98 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr5_+_37903790 | 2.93 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr24_-_38110779 | 2.78 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr10_+_15255198 | 2.68 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr25_+_31267268 | 2.60 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr5_+_32206378 | 2.55 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr25_+_3327071 | 2.50 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr16_-_28658341 | 2.49 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr21_+_45841731 | 2.45 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr10_-_34916208 | 2.43 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr21_+_6751405 | 2.31 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr11_+_13224281 | 2.29 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr22_-_20695237 | 2.23 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr5_+_28497956 | 2.21 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr21_+_6751760 | 2.18 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr9_+_8390737 | 2.15 |
ENSDART00000190891
ENSDART00000176877 ENSDART00000144851 ENSDART00000133880 ENSDART00000142233 |
zgc:113984
|
zgc:113984 |
| chr21_+_31150438 | 2.09 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr17_+_51627209 | 2.06 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr13_-_52089003 | 2.05 |
ENSDART00000187600
|
tmem254
|
transmembrane protein 254 |
| chr8_+_24745041 | 2.01 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr25_+_31276842 | 1.98 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr13_-_9886579 | 1.84 |
ENSDART00000101926
|
si:ch211-117n7.7
|
si:ch211-117n7.7 |
| chr22_+_7738966 | 1.82 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr5_-_68333081 | 1.81 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr16_+_47207691 | 1.78 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr23_+_32101202 | 1.74 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr23_+_26026383 | 1.71 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr10_+_33393829 | 1.70 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr24_-_27400017 | 1.69 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr15_+_34934568 | 1.66 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr17_-_4245902 | 1.65 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr20_-_33566640 | 1.62 |
ENSDART00000159729
|
si:dkey-65b13.9
|
si:dkey-65b13.9 |
| chr13_-_29420885 | 1.61 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr25_+_22320738 | 1.57 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr8_+_45334255 | 1.56 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr21_+_31150773 | 1.55 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr4_-_8043839 | 1.54 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr7_+_56577522 | 1.53 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr3_+_3681116 | 1.51 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr9_-_12443726 | 1.49 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr4_+_5249494 | 1.48 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr17_-_12336987 | 1.45 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr5_+_32260502 | 1.44 |
ENSDART00000149020
|
si:ch211-158m24.12
|
si:ch211-158m24.12 |
| chr4_-_5019113 | 1.42 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr16_-_12173554 | 1.42 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr22_+_7439186 | 1.42 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr21_+_28478663 | 1.41 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr17_+_23298928 | 1.41 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr4_-_815871 | 1.38 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr6_+_21001264 | 1.38 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr25_+_36292057 | 1.37 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr6_+_32882821 | 1.37 |
ENSDART00000075780
|
ghrhrb
|
growth hormone releasing hormone receptor b |
| chr15_+_44366556 | 1.36 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr10_+_6884627 | 1.32 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr8_-_50888806 | 1.30 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr9_+_34127005 | 1.28 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr7_+_59020972 | 1.27 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr2_-_21082695 | 1.24 |
ENSDART00000032502
|
nebl
|
nebulette |
| chr10_+_21867307 | 1.24 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr5_-_1203455 | 1.23 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr12_+_18578597 | 1.19 |
ENSDART00000134944
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr4_+_18804317 | 1.16 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr13_-_8692860 | 1.16 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr18_+_7073130 | 1.14 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr7_+_23515966 | 1.12 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr9_+_54039006 | 1.12 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr16_-_12173399 | 1.12 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr15_+_29727799 | 1.10 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr12_+_47081783 | 1.09 |
ENSDART00000158568
|
mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr17_-_2834764 | 1.09 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr10_+_21722892 | 1.08 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr19_-_9867001 | 1.07 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr9_+_48761455 | 1.07 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr23_-_41965557 | 1.06 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr1_+_35985813 | 1.05 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr10_-_26738209 | 1.05 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr15_-_33925851 | 1.04 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr12_+_16168342 | 1.04 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr3_-_13461056 | 1.03 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr21_+_45502621 | 1.03 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr13_-_31452516 | 1.01 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr19_-_5103141 | 1.00 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr8_+_25767610 | 0.99 |
ENSDART00000062406
|
cacna1sb
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, b |
| chr2_-_55797318 | 0.99 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr7_+_20471315 | 0.99 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr4_+_4849789 | 0.96 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr3_+_16722014 | 0.96 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr13_-_39736938 | 0.95 |
ENSDART00000141645
|
zgc:171482
|
zgc:171482 |
| chr2_+_42871831 | 0.93 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr13_+_35474235 | 0.92 |
ENSDART00000181927
|
mkks
|
McKusick-Kaufman syndrome |
| chr15_+_34592215 | 0.92 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr19_+_5480327 | 0.92 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr8_-_30979494 | 0.91 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr23_-_14766902 | 0.91 |
ENSDART00000168113
|
gss
|
glutathione synthetase |
| chr4_-_2219705 | 0.90 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr12_-_41684729 | 0.89 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr1_+_12295142 | 0.88 |
ENSDART00000158595
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr13_+_8693410 | 0.88 |
ENSDART00000138448
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr9_+_18829360 | 0.87 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr17_-_40956035 | 0.86 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr14_-_49063157 | 0.86 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr2_-_42871286 | 0.85 |
ENSDART00000087823
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr23_+_18527028 | 0.85 |
ENSDART00000190263
ENSDART00000104545 |
avpr2ab
|
arginine vasopressin receptor 2a, duplicate b |
| chr20_-_7583486 | 0.83 |
ENSDART00000144729
|
usp24
|
ubiquitin specific peptidase 24 |
| chr9_-_3866707 | 0.82 |
ENSDART00000010378
|
myo3b
|
myosin IIIB |
| chr9_-_35334642 | 0.82 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr1_+_17676745 | 0.81 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr11_+_41540862 | 0.81 |
ENSDART00000173210
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr6_+_7421898 | 0.81 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr23_-_24825863 | 0.80 |
ENSDART00000112493
|
syt6a
|
synaptotagmin VIa |
| chr17_-_6613458 | 0.80 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr10_-_20706109 | 0.79 |
ENSDART00000124751
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr8_-_53044300 | 0.78 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr3_+_62000822 | 0.78 |
ENSDART00000106680
|
rai1
|
retinoic acid induced 1 |
| chr24_+_40860320 | 0.78 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr1_-_50859053 | 0.77 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr18_+_45958375 | 0.77 |
ENSDART00000108636
|
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr4_-_948776 | 0.76 |
ENSDART00000023483
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr25_-_19420949 | 0.76 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr23_+_8989168 | 0.76 |
ENSDART00000034380
|
xkr7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr19_-_5103313 | 0.75 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr20_-_52902693 | 0.75 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr2_+_50608099 | 0.75 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr3_+_46459540 | 0.74 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr5_+_42097608 | 0.74 |
ENSDART00000014404
|
p2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr3_+_21225750 | 0.74 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr7_+_27898091 | 0.74 |
ENSDART00000186451
ENSDART00000173510 |
tub
|
tubby bipartite transcription factor |
| chr3_-_6519691 | 0.73 |
ENSDART00000165273
ENSDART00000179882 ENSDART00000172292 |
GGA3 (1 of many)
|
si:ch73-157i16.3 |
| chr15_+_45544589 | 0.73 |
ENSDART00000055978
|
LO018197.1
|
|
| chr3_+_40164129 | 0.72 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr20_+_715739 | 0.72 |
ENSDART00000136768
|
myo6a
|
myosin VIa |
| chr16_+_32029090 | 0.71 |
ENSDART00000041054
|
tmc4
|
transmembrane channel-like 4 |
| chr2_+_47708853 | 0.71 |
ENSDART00000124307
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr1_+_49668423 | 0.71 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr6_+_39905021 | 0.70 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr11_-_29737088 | 0.70 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr16_-_41646164 | 0.70 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr14_-_858985 | 0.70 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr17_-_22573311 | 0.70 |
ENSDART00000141523
ENSDART00000140022 ENSDART00000079390 ENSDART00000188644 |
exo1
|
exonuclease 1 |
| chr16_+_11660839 | 0.69 |
ENSDART00000193911
ENSDART00000143683 |
si:dkey-250k15.10
|
si:dkey-250k15.10 |
| chr24_-_7697274 | 0.69 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr7_-_7692723 | 0.69 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr23_+_28809002 | 0.68 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr11_+_30057762 | 0.68 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr13_-_11644806 | 0.68 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr5_-_20491198 | 0.68 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr13_+_6342410 | 0.68 |
ENSDART00000065228
|
csmd1a
|
CUB and Sushi multiple domains 1a |
| chr7_-_8712148 | 0.67 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr8_+_7097929 | 0.67 |
ENSDART00000188955
ENSDART00000184772 ENSDART00000109581 |
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr23_-_37113215 | 0.67 |
ENSDART00000146835
|
zgc:193690
|
zgc:193690 |
| chr17_+_16564921 | 0.66 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr13_+_35528607 | 0.65 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr16_-_13281380 | 0.65 |
ENSDART00000103882
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr17_-_36896560 | 0.65 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr15_-_2519640 | 0.64 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr9_-_50000144 | 0.64 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr5_+_12888260 | 0.64 |
ENSDART00000175916
|
gal3st1a
|
galactose-3-O-sulfotransferase 1a |
| chr24_+_5912635 | 0.64 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr16_+_2820340 | 0.63 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr21_-_2814709 | 0.63 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr5_-_12219572 | 0.63 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr14_-_25309058 | 0.62 |
ENSDART00000159569
|
htr4
|
5-hydroxytryptamine receptor 4 |
| chr4_+_9669717 | 0.62 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr25_+_34938317 | 0.62 |
ENSDART00000042678
|
vps4a
|
vacuolar protein sorting 4a homolog A (S. cerevisiae) |
| chr12_-_47648538 | 0.61 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr15_-_18162647 | 0.61 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr14_+_36521005 | 0.61 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr18_+_26337869 | 0.60 |
ENSDART00000109257
|
RASGRF1
|
si:ch211-234p18.3 |
| chr22_+_35275468 | 0.60 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr18_+_38908903 | 0.60 |
ENSDART00000159834
|
myo5aa
|
myosin VAa |
| chr20_+_23173710 | 0.59 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr17_+_5793248 | 0.58 |
ENSDART00000153743
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr12_+_16087077 | 0.58 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr11_-_27867024 | 0.58 |
ENSDART00000182136
ENSDART00000187587 |
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr1_-_29139141 | 0.58 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr10_-_14556978 | 0.58 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
| chr12_+_31735159 | 0.57 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr7_+_20629411 | 0.57 |
ENSDART00000173710
|
si:dkey-19b23.15
|
si:dkey-19b23.15 |
| chr10_+_45071603 | 0.57 |
ENSDART00000186505
ENSDART00000157573 |
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr17_-_200316 | 0.56 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr17_+_28533102 | 0.56 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr19_+_46158078 | 0.56 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr3_-_23407720 | 0.56 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr24_-_31843173 | 0.55 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr10_+_25219728 | 0.55 |
ENSDART00000193829
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr16_+_4695075 | 0.55 |
ENSDART00000039054
|
mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr8_+_635704 | 0.55 |
ENSDART00000130358
|
csgalnact1b
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1b |
| chr21_+_17956024 | 0.55 |
ENSDART00000142468
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr20_-_46128590 | 0.55 |
ENSDART00000123744
|
taar1b
|
trace amine associated receptor 1b |
| chr7_-_7692992 | 0.54 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr20_+_39250673 | 0.54 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr4_+_61995745 | 0.54 |
ENSDART00000171539
|
CT990567.1
|
|
| chr6_-_15492030 | 0.54 |
ENSDART00000156141
ENSDART00000183992 |
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax7a+pax7b+prop1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.4 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.7 | 4.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 1.6 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.5 | 4.3 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.4 | 1.8 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 1.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.4 | 1.8 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.4 | 1.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.3 | 1.7 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.3 | 3.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 1.0 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 1.6 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.3 | 1.4 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.2 | 0.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 0.6 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.2 | 0.8 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.2 | 3.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.6 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.2 | 0.6 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.2 | 0.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 0.5 | GO:0044406 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.2 | 0.5 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.7 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.6 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 1.5 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.1 | 0.4 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.5 | GO:0044060 | regulation of endocrine process(GO:0044060) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) endocrine hormone secretion(GO:0060986) |
| 0.1 | 0.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 1.3 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 1.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.8 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.1 | 0.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.3 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.6 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 1.8 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.8 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 1.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 1.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.3 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.8 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 1.6 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 1.1 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 0.7 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 0.6 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.5 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 1.3 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.8 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.7 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.9 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 1.1 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 2.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 0.7 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.2 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.3 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.4 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.3 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) positive regulation of voltage-gated calcium channel activity(GO:1901387) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.1 | 2.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 2.6 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.2 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.1 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 2.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.5 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 1.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.4 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.6 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.3 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 1.8 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.5 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 1.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.4 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.8 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.6 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.7 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.2 | GO:0000097 | sulfur amino acid biosynthetic process(GO:0000097) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) lipid glycosylation(GO:0030259) |
| 0.0 | 0.6 | GO:0045761 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 1.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.3 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.4 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.9 | GO:0042593 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 1.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 1.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 2.5 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 1.0 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 1.0 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 1.3 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.4 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0090280 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 2.7 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 1.1 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 3.5 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.3 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0060078 | regulation of postsynaptic membrane potential(GO:0060078) |
| 0.0 | 1.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.0 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.4 | 1.7 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 1.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 0.7 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 2.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.2 | 0.9 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.8 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.4 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.1 | 1.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.4 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 2.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 1.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 4.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.6 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 2.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 1.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.9 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 15.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 3.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.7 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.6 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.8 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 5.0 | GO:0045202 | synapse(GO:0045202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 1.1 | 4.3 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.8 | 3.4 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.8 | 2.5 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.7 | 2.7 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.6 | 4.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 1.6 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.5 | 1.5 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.4 | 1.8 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 1.1 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.4 | 2.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 2.5 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.3 | 1.3 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.3 | 3.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 0.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.3 | 0.9 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.2 | 0.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 0.7 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.2 | 1.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 0.8 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.2 | 1.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 0.9 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 1.2 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 1.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.7 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.6 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.3 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.8 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.7 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 2.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 2.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 2.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.5 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 2.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 2.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 4.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.0 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 1.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.1 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 1.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 1.8 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.3 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.3 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0004527 | exonuclease activity(GO:0004527) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 2.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 1.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 0.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.3 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |