Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
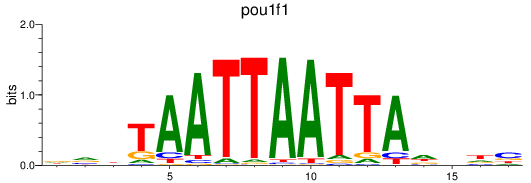
Results for pou1f1
Z-value: 0.63
Transcription factors associated with pou1f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou1f1
|
ENSDARG00000058924 | POU class 1 homeobox 1 |
|
pou1f1
|
ENSDARG00000110816 | POU class 1 homeobox 1 |
|
pou1f1
|
ENSDARG00000115109 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou1f1 | dr11_v1_chr9_-_19161982_19161982 | -0.10 | 3.2e-01 | Click! |
Activity profile of pou1f1 motif
Sorted Z-values of pou1f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_17712393 | 17.90 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr13_+_24279021 | 7.61 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr3_+_26145013 | 6.17 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr19_+_46158078 | 5.96 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr7_+_59020972 | 5.57 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr5_-_44829719 | 5.20 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr25_+_14087045 | 4.77 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr13_+_9432501 | 4.69 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr23_+_26026383 | 4.31 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr22_-_31060579 | 3.55 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr15_-_20939579 | 3.38 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr1_+_5402476 | 3.03 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr6_+_40714811 | 3.03 |
ENSDART00000153868
|
ccdc36
|
coiled-coil domain containing 36 |
| chr3_-_59297532 | 2.89 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr5_-_71705191 | 2.84 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr5_+_71802014 | 2.79 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr22_-_24818066 | 2.69 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr20_-_40755614 | 2.63 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr16_+_12836143 | 2.58 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr5_+_33498253 | 2.42 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr9_-_22099536 | 2.39 |
ENSDART00000101923
|
CR391987.1
|
|
| chr4_-_948776 | 2.32 |
ENSDART00000023483
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr5_-_1203455 | 2.31 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr13_-_31622195 | 2.30 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr8_-_53044300 | 2.22 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr16_+_51326237 | 2.09 |
ENSDART00000168525
|
CU469384.1
|
|
| chr4_+_22480169 | 2.07 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr13_-_12602920 | 2.06 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr22_-_24738188 | 2.00 |
ENSDART00000050238
|
vtg1
|
vitellogenin 1 |
| chr2_+_20496223 | 1.99 |
ENSDART00000157931
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr5_-_41494831 | 1.90 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr18_+_39487486 | 1.83 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr19_+_31873308 | 1.66 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr9_+_54290896 | 1.64 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr23_-_18707418 | 1.61 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr9_+_48761455 | 1.60 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr17_-_30702411 | 1.59 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr2_+_36004381 | 1.58 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr16_-_38118003 | 1.52 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr19_+_43780970 | 1.50 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr7_+_21272833 | 1.48 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr2_+_20605925 | 1.48 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr2_+_49799470 | 1.45 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr10_+_21867307 | 1.44 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr17_+_30591287 | 1.43 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr8_+_17987215 | 1.42 |
ENSDART00000113605
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr7_+_51774502 | 1.42 |
ENSDART00000007767
|
got2b
|
glutamic-oxaloacetic transaminase 2b, mitochondrial |
| chr7_-_7692723 | 1.41 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr22_+_1556948 | 1.41 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr23_+_32101202 | 1.39 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr18_+_27821856 | 1.39 |
ENSDART00000131712
|
si:ch211-222m18.4
|
si:ch211-222m18.4 |
| chr23_+_4709607 | 1.36 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr14_+_36521005 | 1.35 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr5_+_2815021 | 1.34 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr9_+_28598577 | 1.34 |
ENSDART00000142623
ENSDART00000135947 |
si:ch73-7i4.1
|
si:ch73-7i4.1 |
| chr7_+_4694762 | 1.34 |
ENSDART00000132862
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr25_+_22320738 | 1.31 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr23_-_41965557 | 1.31 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr17_+_15559046 | 1.30 |
ENSDART00000187126
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr13_-_39736938 | 1.29 |
ENSDART00000141645
|
zgc:171482
|
zgc:171482 |
| chr16_-_25608453 | 1.29 |
ENSDART00000140140
|
zgc:110410
|
zgc:110410 |
| chr25_+_5972690 | 1.28 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr13_-_9886579 | 1.27 |
ENSDART00000101926
|
si:ch211-117n7.7
|
si:ch211-117n7.7 |
| chr15_+_9327252 | 1.26 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr8_+_17987778 | 1.26 |
ENSDART00000133429
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr23_-_19140781 | 1.25 |
ENSDART00000143580
|
si:ch73-381f5.2
|
si:ch73-381f5.2 |
| chr7_+_49800478 | 1.25 |
ENSDART00000174254
|
PAMR1 (1 of many)
|
si:ch211-102l7.3 |
| chr25_-_4525081 | 1.22 |
ENSDART00000184347
|
pidd1
|
p53-induced death domain protein 1 |
| chr22_+_18469004 | 1.21 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr12_-_28363111 | 1.21 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr7_-_7692992 | 1.20 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr17_+_2162916 | 1.20 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr17_+_52524452 | 1.19 |
ENSDART00000016614
|
dlst
|
dihydrolipoamide S-succinyltransferase |
| chr24_+_9475809 | 1.19 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr8_+_24745041 | 1.19 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr2_+_14992879 | 1.19 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr4_+_19700308 | 1.18 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr22_-_9183944 | 1.17 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr2_-_16216568 | 1.17 |
ENSDART00000173758
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr5_-_54672763 | 1.17 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr12_+_20587179 | 1.16 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr20_+_4060839 | 1.15 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr6_+_6797520 | 1.14 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr4_+_25654686 | 1.14 |
ENSDART00000100714
|
acot16
|
acyl-CoA thioesterase 16 |
| chr20_-_7583486 | 1.14 |
ENSDART00000144729
|
usp24
|
ubiquitin specific peptidase 24 |
| chr7_+_4694924 | 1.14 |
ENSDART00000144873
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr14_+_32430982 | 1.14 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr22_+_14836291 | 1.14 |
ENSDART00000122740
|
gtpbp1l
|
GTP binding protein 1, like |
| chr7_+_24573721 | 1.12 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr25_-_37186894 | 1.11 |
ENSDART00000191647
ENSDART00000182095 |
tdrd12
|
tudor domain containing 12 |
| chr6_+_3809346 | 1.11 |
ENSDART00000185105
|
FO704755.1
|
|
| chr17_-_10122204 | 1.11 |
ENSDART00000160751
|
BX088587.1
|
|
| chr9_-_394088 | 1.11 |
ENSDART00000169014
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr7_-_66868543 | 1.10 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr8_-_53044089 | 1.10 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr2_-_7845110 | 1.09 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr9_+_11281969 | 1.09 |
ENSDART00000110691
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr17_-_27048537 | 1.08 |
ENSDART00000050018
ENSDART00000193861 |
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr2_+_36007449 | 1.07 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr17_+_45602640 | 1.07 |
ENSDART00000153822
|
si:ch211-202f3.4
|
si:ch211-202f3.4 |
| chr14_+_46313396 | 1.07 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr15_-_46821568 | 1.06 |
ENSDART00000148525
|
chrd
|
chordin |
| chr1_+_27690 | 1.06 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr13_+_35528607 | 1.05 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr22_-_14247276 | 1.05 |
ENSDART00000033332
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr24_+_21621654 | 1.03 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr23_-_42232124 | 1.02 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr8_-_13046089 | 1.02 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr3_+_52545014 | 1.01 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr2_-_10877765 | 1.00 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr24_-_21847945 | 0.99 |
ENSDART00000189097
|
CR352265.4
|
|
| chr7_+_49800849 | 0.99 |
ENSDART00000164838
ENSDART00000187888 |
PAMR1 (1 of many)
|
si:ch211-102l7.3 |
| chr2_+_20406399 | 0.98 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr12_-_6880694 | 0.98 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr10_+_36197968 | 0.98 |
ENSDART00000114102
|
COA4
|
si:ch211-215a9.5 |
| chr15_-_39955785 | 0.98 |
ENSDART00000154556
|
msh5
|
mutS homolog 5 |
| chr13_+_31211050 | 0.97 |
ENSDART00000111705
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr7_-_17412559 | 0.97 |
ENSDART00000163020
|
nitr3a
|
novel immune-type receptor 3a |
| chr14_+_14806692 | 0.96 |
ENSDART00000193050
|
fhdc2
|
FH2 domain containing 2 |
| chr9_+_29548195 | 0.96 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr10_+_5645887 | 0.95 |
ENSDART00000171426
|
pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr6_-_28980756 | 0.94 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr15_-_40157331 | 0.94 |
ENSDART00000187958
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr6_-_58764672 | 0.94 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr15_-_40157513 | 0.93 |
ENSDART00000184014
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr20_+_10723292 | 0.93 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr8_-_2434282 | 0.93 |
ENSDART00000137262
ENSDART00000134044 |
vdac3
|
voltage-dependent anion channel 3 |
| chr3_+_23029934 | 0.92 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr17_+_12285285 | 0.92 |
ENSDART00000154336
|
pimr174
|
Pim proto-oncogene, serine/threonine kinase, related 174 |
| chr3_-_39180048 | 0.92 |
ENSDART00000049720
|
cdk21
|
cyclin-dependent kinase 21 |
| chr6_-_52788213 | 0.92 |
ENSDART00000179880
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr5_+_12888260 | 0.92 |
ENSDART00000175916
|
gal3st1a
|
galactose-3-O-sulfotransferase 1a |
| chr8_-_45729248 | 0.92 |
ENSDART00000187736
|
BX088696.1
|
|
| chr6_-_16948040 | 0.91 |
ENSDART00000156433
|
pimr30
|
Pim proto-oncogene, serine/threonine kinase, related 30 |
| chr3_+_23029484 | 0.90 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr19_-_20403845 | 0.90 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr8_-_45729758 | 0.90 |
ENSDART00000189329
|
BX088696.1
|
|
| chr12_-_48006835 | 0.90 |
ENSDART00000108989
|
adamts14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr1_-_42289704 | 0.89 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr17_+_20589553 | 0.89 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr7_-_17425535 | 0.89 |
ENSDART00000098053
ENSDART00000161873 |
nitr3c
|
novel immune-type receptor 3c |
| chr16_+_21016820 | 0.89 |
ENSDART00000191861
|
BX571678.1
|
|
| chr20_-_43663494 | 0.89 |
ENSDART00000144564
|
BX470188.1
|
|
| chr23_-_16484383 | 0.89 |
ENSDART00000187839
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr17_-_48944465 | 0.89 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr3_-_12026741 | 0.88 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr9_+_7724152 | 0.88 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr1_+_49668423 | 0.87 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr11_-_40742424 | 0.87 |
ENSDART00000173399
ENSDART00000021369 |
tas1r3
|
taste receptor, type 1, member 3 |
| chr6_-_17179637 | 0.86 |
ENSDART00000153710
|
pimr19
|
Pim proto-oncogene, serine/threonine kinase, related 19 |
| chr6_-_16804001 | 0.86 |
ENSDART00000155398
|
pimr40
|
Pim proto-oncogene, serine/threonine kinase, related 40 |
| chr22_+_14836040 | 0.86 |
ENSDART00000180951
|
gtpbp1l
|
GTP binding protein 1, like |
| chr6_-_17266221 | 0.85 |
ENSDART00000155448
|
pimr18
|
Pim proto-oncogene, serine/threonine kinase, related 18 |
| chr3_-_10749691 | 0.85 |
ENSDART00000183088
|
CU210890.1
|
|
| chr6_-_16735402 | 0.84 |
ENSDART00000154216
|
pimr44
|
Pim proto-oncogene, serine/threonine kinase, related 44 |
| chr18_+_7073130 | 0.84 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr6_-_17112151 | 0.84 |
ENSDART00000153546
|
pimr17
|
Pim proto-oncogene, serine/threonine kinase, related 17 |
| chr6_+_612594 | 0.84 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr16_-_27566552 | 0.83 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr10_+_26747755 | 0.83 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr6_-_59271270 | 0.83 |
ENSDART00000126870
|
ndufa4l2b
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2b |
| chr7_-_71531846 | 0.83 |
ENSDART00000111797
|
acer2
|
alkaline ceramidase 2 |
| chr6_-_17052116 | 0.83 |
ENSDART00000156825
|
pimr45
|
Pim proto-oncogene, serine/threonine kinase, related 45 |
| chr22_+_5532003 | 0.83 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr11_-_1935883 | 0.82 |
ENSDART00000172885
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr6_-_16868834 | 0.81 |
ENSDART00000156503
|
pimr41
|
Pim proto-oncogene, serine/threonine kinase, related 41 |
| chr18_+_35407921 | 0.81 |
ENSDART00000057733
|
zgc:153119
|
zgc:153119 |
| chr17_-_29224908 | 0.81 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr22_+_23359369 | 0.80 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr19_-_44955710 | 0.80 |
ENSDART00000165246
|
CSMD3 (1 of many)
|
si:ch211-233f16.1 |
| chr11_-_10456387 | 0.80 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr2_+_19522082 | 0.80 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr8_-_25422186 | 0.80 |
ENSDART00000113492
ENSDART00000131736 |
kcnq2a
|
potassium voltage-gated channel, KQT-like subfamily, member 2a |
| chr7_+_4702269 | 0.80 |
ENSDART00000139880
|
si:ch211-225k7.4
|
si:ch211-225k7.4 |
| chr13_-_38039871 | 0.80 |
ENSDART00000140645
|
CR456624.1
|
|
| chr10_-_45029041 | 0.79 |
ENSDART00000167878
|
polm
|
polymerase (DNA directed), mu |
| chr9_+_1039215 | 0.79 |
ENSDART00000192186
|
DOCK9 (1 of many)
|
dedicator of cytokinesis 9 |
| chr2_+_19578446 | 0.79 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr17_-_46817295 | 0.78 |
ENSDART00000155904
|
pimr24
|
Pim proto-oncogene, serine/threonine kinase, related 24 |
| chr25_+_36345867 | 0.78 |
ENSDART00000186527
|
zgc:165555
|
zgc:165555 |
| chr21_-_25522510 | 0.77 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr17_+_2727807 | 0.77 |
ENSDART00000178759
|
kcnk10b
|
potassium channel, subfamily K, member 10b |
| chr12_-_3773869 | 0.77 |
ENSDART00000092983
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr9_-_7652792 | 0.77 |
ENSDART00000137957
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr3_+_40164129 | 0.77 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr10_+_11261576 | 0.76 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr5_+_6672870 | 0.76 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr12_+_47698356 | 0.76 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr1_-_46244523 | 0.76 |
ENSDART00000143908
|
si:ch211-138g9.3
|
si:ch211-138g9.3 |
| chr15_+_37965953 | 0.75 |
ENSDART00000154192
|
si:dkey-238d18.10
|
si:dkey-238d18.10 |
| chr11_-_29737088 | 0.75 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr8_+_10001805 | 0.74 |
ENSDART00000132894
|
si:dkey-8e10.2
|
si:dkey-8e10.2 |
| chr7_+_71664624 | 0.74 |
ENSDART00000170273
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr18_-_3473081 | 0.72 |
ENSDART00000174975
|
myo7aa
|
myosin VIIAa |
| chr2_+_42330828 | 0.72 |
ENSDART00000134802
ENSDART00000193783 |
ftr09
|
finTRIM family, member 9 |
| chr19_-_20403318 | 0.72 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr15_-_39945036 | 0.72 |
ENSDART00000192481
|
msh5
|
mutS homolog 5 |
| chr23_-_18024543 | 0.71 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr11_-_2838699 | 0.71 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr10_+_42423318 | 0.71 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou1f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0046351 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 1.5 | 6.2 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 1.2 | 3.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.6 | 2.3 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.5 | 2.6 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.5 | 1.8 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.4 | 1.6 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.4 | 1.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.4 | 3.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.4 | 1.8 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.4 | 2.2 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.4 | 1.4 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.3 | 1.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 2.2 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.3 | 0.8 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.3 | 4.3 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.3 | 1.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.3 | 1.3 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.2 | 1.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.7 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.2 | 1.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 5.6 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.2 | 1.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.2 | 1.0 | GO:0031650 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.2 | 6.0 | GO:0030817 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.2 | 0.7 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.2 | 0.5 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.2 | 1.0 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 1.7 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.6 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 2.0 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 5.3 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 1.6 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.1 | 0.7 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.4 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 1.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.5 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.6 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.9 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.3 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.9 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.9 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 1.0 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.4 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.4 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 7.6 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 1.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.6 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 0.3 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 1.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.6 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 0.4 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 3.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 1.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.6 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 2.1 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.1 | 0.9 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 3.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.5 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.9 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.2 | GO:0048618 | embryonic digestive tract morphogenesis(GO:0048557) post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.6 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 1.1 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 1.2 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.3 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 14.8 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 4.4 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 2.7 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 3.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.8 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 2.4 | GO:0007599 | hemostasis(GO:0007599) |
| 0.0 | 1.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 1.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.5 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.5 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 1.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.1 | GO:0016571 | histone methylation(GO:0016571) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.9 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.0 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 2.4 | GO:1901605 | alpha-amino acid metabolic process(GO:1901605) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.2 | GO:0031673 | H zone(GO:0031673) |
| 1.1 | 4.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.5 | 7.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 2.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 2.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 3.0 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 0.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 2.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.6 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 6.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 4.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.7 | 5.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 1.4 | 5.6 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.7 | 4.7 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.6 | 1.8 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.6 | 1.8 | GO:0034618 | arginine binding(GO:0034618) |
| 0.5 | 1.6 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.5 | 2.0 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.5 | 1.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.4 | 1.3 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.4 | 1.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.4 | 1.6 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.4 | 1.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.4 | 2.6 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.4 | 6.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 0.9 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.3 | 4.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 0.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 2.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 0.9 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 0.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 2.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.8 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.5 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 0.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.4 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 3.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.3 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 1.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.6 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.7 | GO:0038187 | pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.6 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.3 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 3.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.2 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.4 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 1.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 4.9 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.8 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 3.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 3.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 15.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 1.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 6.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 3.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 6.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 2.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |