Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
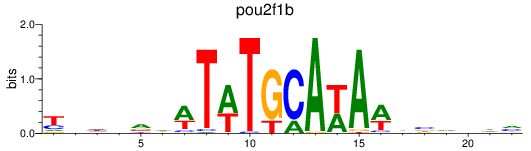
Results for pou2f1b
Z-value: 1.07
Transcription factors associated with pou2f1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f1b
|
ENSDARG00000007996 | POU class 2 homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f1b | dr11_v1_chr9_+_34334156_34334156 | -0.46 | 3.5e-06 | Click! |
Activity profile of pou2f1b motif
Sorted Z-values of pou2f1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_35458190 | 19.22 |
ENSDART00000051313
|
fbp1b
|
fructose-1,6-bisphosphatase 1b |
| chr20_+_10538025 | 12.60 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr24_+_23959487 | 12.27 |
ENSDART00000080487
|
hgd
|
homogentisate 1,2-dioxygenase |
| chr19_+_2835240 | 11.79 |
ENSDART00000190838
|
CDCP1
|
CUB domain containing protein 1 |
| chr24_-_21150100 | 11.40 |
ENSDART00000154249
ENSDART00000193256 |
gramd1c
|
GRAM domain containing 1c |
| chr20_+_10544100 | 10.38 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr1_-_56080112 | 9.94 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr18_-_6742287 | 9.73 |
ENSDART00000140752
|
miox
|
myo-inositol oxygenase |
| chr4_-_8043839 | 8.84 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr1_-_10071422 | 8.59 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr2_-_20120904 | 7.45 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr20_-_35750810 | 7.30 |
ENSDART00000153072
|
adgrf8
|
adhesion G protein-coupled receptor F8 |
| chr2_+_27394798 | 7.01 |
ENSDART00000115071
|
selenop2
|
selenoprotein P2 |
| chr22_-_23706771 | 6.86 |
ENSDART00000159771
|
cfhl1
|
complement factor H like 1 |
| chr12_+_22560067 | 6.86 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr19_-_977849 | 6.68 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr14_-_4145594 | 6.64 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr4_+_5249494 | 6.52 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr12_-_4243268 | 6.41 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr25_-_13029680 | 6.16 |
ENSDART00000166151
|
ccl39.2
|
chemokine (C-C motif) ligand 39, duplicate 2 |
| chr17_-_4231532 | 5.91 |
ENSDART00000019364
|
bmp2a
|
bone morphogenetic protein 2a |
| chr17_+_23311377 | 5.88 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr15_+_14856307 | 5.84 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr3_+_49021079 | 5.58 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr2_-_43635777 | 5.57 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr2_+_37875789 | 5.56 |
ENSDART00000036318
ENSDART00000127679 |
cbln13
|
cerebellin 13 |
| chr16_+_19637384 | 5.53 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr22_-_23668356 | 5.46 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr14_-_46198373 | 5.42 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr18_+_44703343 | 5.41 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr9_-_12443726 | 5.29 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr2_+_31665836 | 5.29 |
ENSDART00000135411
ENSDART00000143914 |
si:ch211-106h4.12
|
si:ch211-106h4.12 |
| chr20_-_49889111 | 5.26 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr17_+_28102487 | 5.21 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr3_+_12816362 | 5.19 |
ENSDART00000163743
ENSDART00000170788 |
cyp2k6
|
cytochrome P450, family 2, subfamily K, polypeptide 6 |
| chr1_-_44638058 | 5.11 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr9_-_34396264 | 4.99 |
ENSDART00000045754
|
grtp1b
|
growth hormone regulated TBC protein 1b |
| chr11_-_34065718 | 4.98 |
ENSDART00000110608
|
col6a1
|
collagen, type VI, alpha 1 |
| chr2_-_21438492 | 4.96 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr2_+_49457449 | 4.96 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr13_-_21672131 | 4.95 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr16_+_38394371 | 4.81 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr25_-_18454016 | 4.72 |
ENSDART00000005877
|
cpa1
|
carboxypeptidase A1 (pancreatic) |
| chr15_+_32711663 | 4.65 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr2_+_42191592 | 4.65 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr15_-_18209672 | 4.62 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr16_-_45288599 | 4.56 |
ENSDART00000044097
|
hpn
|
hepsin |
| chr19_-_27570333 | 4.55 |
ENSDART00000146562
ENSDART00000179060 |
si:dkeyp-46h3.5
si:dkeyp-46h3.8
|
si:dkeyp-46h3.5 si:dkeyp-46h3.8 |
| chr23_+_39695827 | 4.52 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr6_-_58764672 | 4.51 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr7_-_12968689 | 4.45 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr5_+_26799165 | 4.33 |
ENSDART00000145736
|
tcn2
|
transcobalamin II |
| chr3_+_37112693 | 4.31 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr14_-_21064199 | 4.31 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr16_-_42872571 | 4.26 |
ENSDART00000154757
ENSDART00000102345 |
txnipb
|
thioredoxin interacting protein b |
| chr23_+_7692042 | 4.26 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr5_+_35458346 | 4.17 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr20_-_35578435 | 4.12 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr16_-_23346095 | 4.11 |
ENSDART00000160546
|
si:dkey-247k7.2
|
si:dkey-247k7.2 |
| chr3_+_49043917 | 4.06 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr24_-_23758003 | 4.04 |
ENSDART00000178085
|
BX640462.2
|
Danio rerio minichromosome maintenance domain containing 2 (mcmdc2), mRNA. |
| chr5_+_39087364 | 3.97 |
ENSDART00000004286
|
anxa3a
|
annexin A3a |
| chr16_+_50289916 | 3.95 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr7_-_38792543 | 3.83 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr14_+_10461264 | 3.80 |
ENSDART00000081095
|
cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr19_-_340641 | 3.79 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr6_+_9191981 | 3.77 |
ENSDART00000150916
|
zgc:112392
|
zgc:112392 |
| chr17_+_2549503 | 3.76 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr18_-_29925717 | 3.70 |
ENSDART00000099281
|
mhc2dbb
|
major histocompatibility complex class II DBB gene |
| chr15_+_19838458 | 3.69 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr20_-_39735952 | 3.59 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr15_-_37867995 | 3.59 |
ENSDART00000192698
|
si:dkey-238d18.4
|
si:dkey-238d18.4 |
| chr8_-_36287046 | 3.57 |
ENSDART00000162877
|
si:busm1-194e12.11
|
si:busm1-194e12.11 |
| chr20_+_52492192 | 3.51 |
ENSDART00000057986
|
TSTA3 (1 of many)
|
zgc:100864 |
| chr18_-_26715156 | 3.44 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr9_-_33063083 | 3.44 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr18_-_7539166 | 3.42 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr1_+_58332000 | 3.42 |
ENSDART00000145234
|
ggt1l2.1
|
gamma-glutamyltransferase 1 like 2.1 |
| chr25_+_186583 | 3.39 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr13_-_25198025 | 3.38 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr19_-_1948236 | 3.34 |
ENSDART00000163344
|
znrf2a
|
zinc and ring finger 2a |
| chr1_+_7546259 | 3.32 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr17_-_38291065 | 3.31 |
ENSDART00000152056
|
pax9
|
paired box 9 |
| chr20_+_18993452 | 3.29 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr6_-_442163 | 3.27 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr18_-_26715655 | 3.25 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr23_+_46183410 | 3.23 |
ENSDART00000167596
ENSDART00000151149 ENSDART00000150896 |
btr31
|
bloodthirsty-related gene family, member 31 |
| chr22_+_29994093 | 3.22 |
ENSDART00000104778
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr10_+_34001444 | 3.18 |
ENSDART00000149934
|
kl
|
klotho |
| chr10_+_35329751 | 3.15 |
ENSDART00000148043
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr1_-_23274393 | 3.09 |
ENSDART00000147800
ENSDART00000130277 ENSDART00000054340 ENSDART00000054338 |
rpl9
|
ribosomal protein L9 |
| chr21_+_8533533 | 3.09 |
ENSDART00000077924
|
FO834888.1
|
|
| chr12_-_20584413 | 3.08 |
ENSDART00000170923
|
FP885542.2
|
|
| chr17_+_16046314 | 3.08 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr4_-_20181964 | 3.07 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr23_-_27701361 | 3.02 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr15_-_35960250 | 3.02 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr21_-_34926619 | 3.01 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr19_-_27550768 | 3.00 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr21_+_15295685 | 2.99 |
ENSDART00000135603
|
BX901878.1
|
|
| chr5_-_50992690 | 2.97 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr23_-_13875252 | 2.97 |
ENSDART00000104834
ENSDART00000193807 |
g6pd
|
glucose-6-phosphate dehydrogenase |
| chr11_-_29658396 | 2.97 |
ENSDART00000183947
|
rpl22
|
ribosomal protein L22 |
| chr11_+_25278772 | 2.97 |
ENSDART00000188630
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr20_-_15099579 | 2.95 |
ENSDART00000127213
|
fmo5
|
flavin containing monooxygenase 5 |
| chr23_-_37575030 | 2.95 |
ENSDART00000031875
|
tor1l3
|
torsin family 1 like 3 |
| chr1_+_58922027 | 2.94 |
ENSDART00000159479
|
trip10b
|
thyroid hormone receptor interactor 10b |
| chr1_+_9153141 | 2.94 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr1_+_51615672 | 2.92 |
ENSDART00000165117
|
zgc:165656
|
zgc:165656 |
| chr5_-_68244564 | 2.86 |
ENSDART00000169350
|
CABZ01083944.1
|
|
| chr17_+_20576848 | 2.85 |
ENSDART00000183203
|
SAMD8
|
zgc:162183 |
| chr4_+_76705830 | 2.85 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr2_+_38039857 | 2.85 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr1_-_55262763 | 2.85 |
ENSDART00000152769
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr21_+_19635486 | 2.83 |
ENSDART00000185736
|
fgf10a
|
fibroblast growth factor 10a |
| chr19_-_1947403 | 2.83 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr24_+_7322116 | 2.80 |
ENSDART00000005804
|
XRCC2
|
X-ray repair cross complementing 2 |
| chr3_+_4403980 | 2.79 |
ENSDART00000156881
|
CR774195.1
|
|
| chr11_-_21404044 | 2.78 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr8_+_24745041 | 2.77 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr7_+_20344222 | 2.75 |
ENSDART00000141186
ENSDART00000139274 |
ponzr1
|
plac8 onzin related protein 1 |
| chr4_+_66853199 | 2.73 |
ENSDART00000170223
|
si:dkey-29m1.2
|
si:dkey-29m1.2 |
| chr11_-_26576754 | 2.72 |
ENSDART00000191733
ENSDART00000002846 |
st3gal8
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 8 |
| chr6_+_8630355 | 2.72 |
ENSDART00000161749
ENSDART00000193976 |
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr22_+_14836291 | 2.71 |
ENSDART00000122740
|
gtpbp1l
|
GTP binding protein 1, like |
| chr8_-_16515127 | 2.70 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr4_-_8035520 | 2.70 |
ENSDART00000146146
|
si:ch211-240l19.7
|
si:ch211-240l19.7 |
| chr17_-_2036850 | 2.67 |
ENSDART00000186048
ENSDART00000188838 ENSDART00000186423 |
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr4_-_5239904 | 2.64 |
ENSDART00000190664
|
ada2b
|
adenosine deaminase 2b |
| chr19_-_7690975 | 2.64 |
ENSDART00000151384
|
si:dkey-204a24.10
|
si:dkey-204a24.10 |
| chr17_-_6618574 | 2.62 |
ENSDART00000184486
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr10_+_38643304 | 2.62 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr22_-_10121880 | 2.62 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr25_-_13050959 | 2.57 |
ENSDART00000169041
|
ccl35.1
|
chemokine (C-C motif) ligand 35, duplicate 1 |
| chr24_-_12770357 | 2.54 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr6_-_19270484 | 2.54 |
ENSDART00000186894
ENSDART00000188709 |
zgc:174863
|
zgc:174863 |
| chr20_-_35470891 | 2.53 |
ENSDART00000152993
ENSDART00000016090 |
pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr6_-_38930726 | 2.53 |
ENSDART00000154151
|
hdac7b
|
histone deacetylase 7b |
| chr15_-_37767901 | 2.53 |
ENSDART00000154763
|
si:dkey-42l23.4
|
si:dkey-42l23.4 |
| chr25_+_29474982 | 2.53 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr4_-_5240128 | 2.52 |
ENSDART00000144048
|
ada2b
|
adenosine deaminase 2b |
| chr4_+_5341592 | 2.52 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr14_-_30390145 | 2.48 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr18_+_38192499 | 2.48 |
ENSDART00000191849
|
nucb2b
|
nucleobindin 2b |
| chr1_+_32051581 | 2.46 |
ENSDART00000146602
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr11_-_21404358 | 2.43 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr20_-_29052391 | 2.42 |
ENSDART00000193482
ENSDART00000017090 |
thbs1b
|
thrombospondin 1b |
| chr23_+_32011768 | 2.40 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr17_+_22530820 | 2.40 |
ENSDART00000089916
|
edaradd
|
EDAR-associated death domain |
| chr13_+_18545819 | 2.39 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr12_-_17655683 | 2.39 |
ENSDART00000066411
|
dlgap5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr15_-_23814330 | 2.38 |
ENSDART00000153843
|
si:ch211-167j9.5
|
si:ch211-167j9.5 |
| chr1_+_32054159 | 2.35 |
ENSDART00000181442
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr23_+_43809484 | 2.33 |
ENSDART00000149534
|
si:ch211-149b19.3
|
si:ch211-149b19.3 |
| chr18_+_39060687 | 2.32 |
ENSDART00000098729
ENSDART00000136367 |
zgc:171509
|
zgc:171509 |
| chr6_-_55399214 | 2.29 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr20_-_41992878 | 2.28 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr8_-_39884359 | 2.27 |
ENSDART00000131372
|
mlec
|
malectin |
| chr23_-_33775145 | 2.26 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr3_-_45326897 | 2.26 |
ENSDART00000086417
|
il21r.2
|
interleukin 21 receptor, tandem duplicate 2 |
| chr21_+_76739 | 2.24 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr15_+_32711172 | 2.21 |
ENSDART00000163936
ENSDART00000168135 |
postnb
|
periostin, osteoblast specific factor b |
| chr16_-_9802449 | 2.21 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr1_-_56556326 | 2.19 |
ENSDART00000188760
|
CABZ01059408.1
|
|
| chr6_-_53160459 | 2.18 |
ENSDART00000128127
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr18_-_3166726 | 2.17 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr9_-_33062891 | 2.17 |
ENSDART00000161182
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr25_+_22320738 | 2.16 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr20_+_4221978 | 2.14 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr18_+_19648275 | 2.11 |
ENSDART00000100569
|
smad6b
|
SMAD family member 6b |
| chr2_+_6253246 | 2.09 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr15_-_43327911 | 2.09 |
ENSDART00000077386
|
prss16
|
protease, serine, 16 (thymus) |
| chr10_-_40819022 | 2.08 |
ENSDART00000076316
ENSDART00000141329 |
zgc:113625
|
zgc:113625 |
| chr9_-_38579758 | 2.07 |
ENSDART00000131738
|
si:dkey-101k6.5
|
si:dkey-101k6.5 |
| chr2_+_43204919 | 2.06 |
ENSDART00000160077
ENSDART00000018729 ENSDART00000129134 ENSDART00000056402 |
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr16_-_50229193 | 2.06 |
ENSDART00000161782
ENSDART00000010081 |
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr3_+_29476085 | 2.03 |
ENSDART00000184495
ENSDART00000181058 |
fam83fa
|
family with sequence similarity 83, member Fa |
| chr1_-_39859626 | 2.01 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr14_-_7409364 | 2.01 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr5_-_1487256 | 2.00 |
ENSDART00000149599
ENSDART00000148411 ENSDART00000092087 ENSDART00000148464 |
golga2
|
golgin A2 |
| chr17_-_6641535 | 2.00 |
ENSDART00000154540
ENSDART00000180384 |
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr10_+_26667475 | 1.99 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr5_+_50879545 | 1.96 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr7_-_51102479 | 1.96 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr4_+_73215536 | 1.95 |
ENSDART00000174290
|
LO018260.2
|
Danio rerio protein NLRC3-like (LOC101883187), mRNA. |
| chr3_+_7771420 | 1.94 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr21_+_25625026 | 1.93 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr16_+_21426524 | 1.92 |
ENSDART00000182869
|
gsdmeb
|
gasdermin Eb |
| chr16_+_40508882 | 1.91 |
ENSDART00000126129
|
nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr24_+_20372497 | 1.91 |
ENSDART00000132340
ENSDART00000139989 ENSDART00000168751 ENSDART00000192909 |
plcd1a
|
phospholipase C, delta 1a |
| chr13_+_30228077 | 1.91 |
ENSDART00000100813
ENSDART00000147729 ENSDART00000133404 |
rps24
|
ribosomal protein S24 |
| chr10_+_29771256 | 1.90 |
ENSDART00000193195
|
hyou1
|
hypoxia up-regulated 1 |
| chr12_-_10381282 | 1.90 |
ENSDART00000152788
|
mki67
|
marker of proliferation Ki-67 |
| chr17_-_21884323 | 1.90 |
ENSDART00000155574
|
cuzd1.1
|
CUB and zona pellucida-like domains 1, tandem duplicate 1 |
| chr14_+_36886950 | 1.90 |
ENSDART00000183719
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr11_+_29975830 | 1.88 |
ENSDART00000148929
|
si:ch73-226l13.2
|
si:ch73-226l13.2 |
| chr21_+_45685757 | 1.87 |
ENSDART00000160530
|
sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr5_+_32914321 | 1.86 |
ENSDART00000051336
|
cyr61l1
|
cysteine-rich, angiogenic inducer 61 like 1 |
| chr15_-_14038227 | 1.86 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr10_+_31809226 | 1.86 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.2 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 2.5 | 7.5 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.7 | 5.2 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 1.6 | 4.8 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 1.5 | 4.6 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 1.4 | 4.3 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 1.2 | 7.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.2 | 5.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 1.0 | 5.2 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 1.0 | 12.3 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.9 | 2.8 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.9 | 4.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.8 | 4.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.8 | 4.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.7 | 9.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.7 | 1.5 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) myoblast migration(GO:0051451) |
| 0.7 | 4.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.7 | 2.8 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.7 | 2.0 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.7 | 2.0 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.6 | 3.7 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.6 | 8.6 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.6 | 3.0 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.6 | 2.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.6 | 4.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.5 | 2.2 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.5 | 3.7 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.5 | 5.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.5 | 2.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.5 | 4.5 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.5 | 1.5 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.5 | 6.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.5 | 1.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.5 | 1.8 | GO:0090076 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.5 | 1.4 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.4 | 1.3 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.4 | 1.8 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.4 | 2.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.4 | 1.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 3.4 | GO:0031179 | peptide modification(GO:0031179) |
| 0.4 | 2.5 | GO:0090467 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.4 | 3.3 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.4 | 1.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.4 | 1.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.4 | 6.9 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.4 | 4.9 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.4 | 1.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.4 | 6.6 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.4 | 7.4 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.3 | 2.0 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.3 | 4.0 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.3 | 6.7 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 | 2.0 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.3 | 1.3 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.3 | 0.9 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.3 | 1.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.3 | 23.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.3 | 2.8 | GO:0072078 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.2 | 3.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 5.0 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 2.1 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.2 | 3.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 1.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 0.8 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 1.0 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.2 | 2.2 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 1.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 1.0 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.2 | 4.1 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.2 | 0.6 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.2 | 5.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 1.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 1.8 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.2 | 2.5 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.2 | 3.7 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.2 | 3.3 | GO:0060021 | palate development(GO:0060021) |
| 0.2 | 5.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.2 | 5.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 2.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 7.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 1.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 1.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 3.0 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.2 | 2.1 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 2.8 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.4 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.1 | 1.0 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.1 | 2.0 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 5.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.4 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 8.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 12.2 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 4.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.5 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 2.1 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.2 | GO:0072178 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.1 | 1.0 | GO:0018202 | histone H3-K36 methylation(GO:0010452) peptidyl-histidine modification(GO:0018202) |
| 0.1 | 1.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.0 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 3.2 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 0.7 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 1.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.8 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 1.9 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 1.8 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 5.0 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.1 | 0.9 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 2.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 1.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.2 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 0.5 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.1 | 0.4 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 0.5 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 0.7 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.6 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 0.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.7 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 2.9 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 1.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 3.7 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 5.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 2.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 1.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 3.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.2 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 1.7 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 4.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.7 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 1.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.6 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 1.8 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 1.0 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.9 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 1.0 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.6 | GO:0060969 | nucleosome positioning(GO:0016584) negative regulation of chromatin silencing(GO:0031936) negative regulation of gene silencing(GO:0060969) |
| 0.0 | 6.8 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 5.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.6 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.0 | 0.7 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.1 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.5 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 1.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 1.3 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 2.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 2.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.3 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.6 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 1.8 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 1.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 2.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 3.4 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 2.8 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.3 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 1.3 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 1.4 | GO:0034249 | negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 1.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.0 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 16.6 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 2.3 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.0 | 0.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.4 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 2.8 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.9 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 1.9 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.9 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 0.6 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 1.1 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.7 | GO:0001503 | ossification(GO:0001503) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 2.0 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.6 | 7.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.6 | 1.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 2.0 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.5 | 1.4 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.5 | 3.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 1.8 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 2.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.4 | 1.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 2.0 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 2.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 1.4 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 0.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 4.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 4.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 2.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 1.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 2.5 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 2.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 4.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 5.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 5.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 7.1 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 2.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 8.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.7 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 4.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 4.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 87.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.8 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 2.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 7.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 2.7 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 5.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 9.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 4.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 25.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 5.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 2.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 2.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 1.8 | 5.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.5 | 7.5 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 1.4 | 7.0 | GO:0008430 | selenium binding(GO:0008430) |
| 1.1 | 4.5 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 1.1 | 5.6 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 1.0 | 3.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 1.0 | 4.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.9 | 4.6 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.9 | 6.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.8 | 9.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.8 | 5.5 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.7 | 2.2 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.7 | 3.3 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.6 | 5.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.5 | 1.6 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.5 | 4.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.5 | 6.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.4 | 1.8 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.4 | 1.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 7.3 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.4 | 3.4 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.4 | 3.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.4 | 1.5 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.4 | 2.2 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.4 | 2.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.3 | 3.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 6.7 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.3 | 6.6 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.3 | 4.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.3 | 2.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 4.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.3 | 0.8 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.3 | 4.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 3.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 1.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 5.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.3 | 1.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 6.9 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.3 | 4.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 12.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 5.0 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 2.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 4.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 2.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 24.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 2.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 0.7 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 3.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 1.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 4.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 9.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.2 | 1.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 3.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.9 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 1.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 2.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 8.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 2.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 4.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.0 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 2.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.4 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.7 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 3.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.0 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 1.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 2.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 4.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 3.0 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 8.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.9 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 12.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.7 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 1.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.4 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 3.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 4.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 9.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 2.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.0 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 5.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.6 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 2.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 5.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 2.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 4.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 2.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 10.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 10.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 3.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 7.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 5.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.8 | 8.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.6 | 7.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.5 | 7.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.4 | 5.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.4 | 3.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.3 | 5.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 8.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 2.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 3.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.2 | 2.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 1.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 7.2 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.2 | 3.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 10.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 3.2 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.2 | 3.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 1.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 8.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 3.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 11.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.1 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 4.5 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |