Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
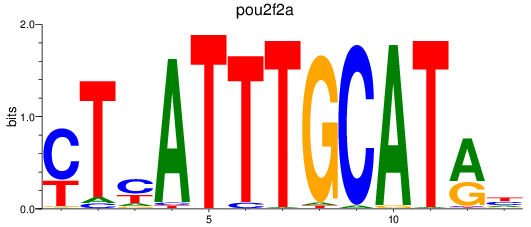
Results for pou2f2a
Z-value: 1.06
Transcription factors associated with pou2f2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f2a
|
ENSDARG00000019658 | POU class 2 homeobox 2a |
|
pou2f2a
|
ENSDARG00000036816 | POU class 2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f2a | dr11_v1_chr16_+_10918252_10918252 | -0.39 | 9.5e-05 | Click! |
Activity profile of pou2f2a motif
Sorted Z-values of pou2f2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_340641 | 12.84 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr24_-_40774028 | 11.11 |
ENSDART00000166578
|
CU633479.2
|
|
| chr17_+_26965351 | 10.07 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr16_+_17714664 | 9.10 |
ENSDART00000149042
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr5_-_63302944 | 8.55 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr9_-_34396264 | 8.28 |
ENSDART00000045754
|
grtp1b
|
growth hormone regulated TBC protein 1b |
| chr19_-_40198478 | 8.25 |
ENSDART00000191736
|
grn2
|
granulin 2 |
| chr16_+_17715243 | 8.06 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr3_+_49021079 | 8.04 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr5_+_26212621 | 7.66 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr25_+_35502552 | 7.63 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr3_-_27646070 | 7.33 |
ENSDART00000122031
ENSDART00000151027 |
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr16_-_24550029 | 7.15 |
ENSDART00000108590
|
si:ch211-79k12.1
|
si:ch211-79k12.1 |
| chr20_+_40150612 | 6.91 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr2_+_38039857 | 6.60 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr17_-_2039511 | 6.40 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr19_+_19775757 | 6.26 |
ENSDART00000164677
|
hoxa3a
|
homeobox A3a |
| chr7_+_66565572 | 6.22 |
ENSDART00000180250
|
tmem176l.3b
|
transmembrane protein 176l.3b |
| chr5_+_58550291 | 6.16 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr3_-_3209432 | 6.00 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr5_+_15202495 | 5.93 |
ENSDART00000144915
|
tbx1
|
T-box 1 |
| chr15_-_29387446 | 5.87 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr5_+_15203421 | 5.83 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr14_-_8080416 | 5.76 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr24_-_25428176 | 5.48 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr21_+_27382893 | 5.47 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr1_+_58332000 | 5.44 |
ENSDART00000145234
|
ggt1l2.1
|
gamma-glutamyltransferase 1 like 2.1 |
| chr24_-_23758003 | 5.30 |
ENSDART00000178085
|
BX640462.2
|
Danio rerio minichromosome maintenance domain containing 2 (mcmdc2), mRNA. |
| chr14_-_15171435 | 5.25 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr17_+_6563307 | 5.23 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr5_-_57686487 | 5.21 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr17_-_25382367 | 5.15 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr6_+_23935045 | 5.15 |
ENSDART00000162993
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr13_-_40411908 | 5.13 |
ENSDART00000057094
ENSDART00000150091 |
nkx2.3
|
NK2 homeobox 3 |
| chr25_+_20188769 | 5.10 |
ENSDART00000142781
|
cald1b
|
caldesmon 1b |
| chr23_+_23232136 | 5.08 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr19_+_14573998 | 4.94 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr20_-_22476255 | 4.76 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr19_+_19988869 | 4.71 |
ENSDART00000151024
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr12_-_4540564 | 4.67 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr17_-_24916889 | 4.58 |
ENSDART00000156157
|
si:ch211-195o20.7
|
si:ch211-195o20.7 |
| chr17_+_15297398 | 4.57 |
ENSDART00000156574
|
si:ch211-270g19.5
|
si:ch211-270g19.5 |
| chr4_-_4612116 | 4.54 |
ENSDART00000130601
|
CABZ01020840.1
|
Danio rerio apoptosis facilitator Bcl-2-like protein 14 (LOC101885512), mRNA. |
| chr19_+_41464870 | 4.40 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr5_-_23749348 | 4.33 |
ENSDART00000140766
|
gbgt1l2
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 2 |
| chr5_-_33287691 | 4.30 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr5_+_26204561 | 4.29 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr11_-_11575070 | 4.21 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr10_+_10351685 | 4.20 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr13_-_6081803 | 4.16 |
ENSDART00000099224
|
dld
|
deltaD |
| chr8_+_20393250 | 4.13 |
ENSDART00000015038
|
zap70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr4_-_9780931 | 4.10 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr24_-_31904924 | 4.08 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr7_+_4931366 | 4.03 |
ENSDART00000172384
ENSDART00000138168 ENSDART00000114543 |
si:dkey-83f18.3
|
si:dkey-83f18.3 |
| chr24_+_39990695 | 3.99 |
ENSDART00000040281
|
BX323854.1
|
|
| chr7_+_4922104 | 3.94 |
ENSDART00000135154
|
si:dkey-28d5.11
|
si:dkey-28d5.11 |
| chr13_-_49444636 | 3.92 |
ENSDART00000136991
|
irf2bp2a
|
interferon regulatory factor 2 binding protein 2a |
| chr12_+_23424108 | 3.80 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr12_+_30586599 | 3.78 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr7_+_3442834 | 3.74 |
ENSDART00000138247
|
si:ch211-285c6.2
|
si:ch211-285c6.2 |
| chr21_+_5993188 | 3.69 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr7_-_51461649 | 3.68 |
ENSDART00000193947
ENSDART00000174328 |
arhgap36
|
Rho GTPase activating protein 36 |
| chr25_+_8407892 | 3.67 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr22_+_980290 | 3.59 |
ENSDART00000065377
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr15_-_29388012 | 3.54 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr24_-_32452355 | 3.50 |
ENSDART00000146511
|
ccdc129
|
coiled-coil domain containing 129 |
| chr2_-_51059691 | 3.48 |
ENSDART00000158131
|
ftr87
|
finTRIM family, member 87 |
| chr2_+_10280645 | 3.46 |
ENSDART00000063996
|
gadd45aa
|
growth arrest and DNA-damage-inducible, alpha, a |
| chr14_-_17622080 | 3.42 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr23_-_20258490 | 3.39 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr23_-_1017605 | 3.33 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr4_-_16406737 | 3.31 |
ENSDART00000013085
|
dcn
|
decorin |
| chr2_-_6482240 | 3.30 |
ENSDART00000132623
|
rgs13
|
regulator of G protein signaling 13 |
| chr7_-_21924372 | 3.23 |
ENSDART00000113652
|
si:dkey-85k7.10
|
si:dkey-85k7.10 |
| chr22_-_3275888 | 3.22 |
ENSDART00000164743
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr25_-_36248053 | 3.21 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T cells 3b |
| chr3_+_29476085 | 3.19 |
ENSDART00000184495
ENSDART00000181058 |
fam83fa
|
family with sequence similarity 83, member Fa |
| chr18_-_7481036 | 3.15 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr1_-_55118745 | 3.15 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr23_+_7692042 | 3.10 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr23_-_1017428 | 3.08 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr21_-_11646878 | 3.08 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr16_+_52999778 | 3.04 |
ENSDART00000011506
|
nkd2a
|
naked cuticle homolog 2a |
| chr14_-_40821411 | 3.03 |
ENSDART00000166621
|
elf1
|
E74-like ETS transcription factor 1 |
| chr7_-_3912905 | 3.01 |
ENSDART00000143864
|
si:dkey-88n24.10
|
si:dkey-88n24.10 |
| chr7_-_8315179 | 2.96 |
ENSDART00000184049
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr22_+_36656680 | 2.96 |
ENSDART00000134031
ENSDART00000056169 |
b3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr14_-_46198373 | 2.94 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr17_-_26867725 | 2.92 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr5_+_54400971 | 2.92 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr13_+_30951155 | 2.91 |
ENSDART00000057469
ENSDART00000162254 |
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr10_+_42169982 | 2.87 |
ENSDART00000190905
|
CU467905.1
|
|
| chr7_+_15736230 | 2.86 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr10_+_8690936 | 2.85 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr7_+_4572727 | 2.84 |
ENSDART00000137830
|
si:dkey-83f18.7
|
si:dkey-83f18.7 |
| chr9_-_42861080 | 2.79 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr9_-_56272465 | 2.77 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr14_-_28001986 | 2.76 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr3_+_2669813 | 2.76 |
ENSDART00000014205
|
CR388047.1
|
|
| chr17_-_26868169 | 2.74 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr6_+_28428329 | 2.73 |
ENSDART00000188056
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr7_+_4795707 | 2.73 |
ENSDART00000142704
|
si:dkey-28d5.4
|
si:dkey-28d5.4 |
| chr10_+_2669405 | 2.71 |
ENSDART00000110202
|
ccl27b
|
chemokine (C-C motif) ligand 27b |
| chr3_-_5228841 | 2.68 |
ENSDART00000092373
ENSDART00000182438 |
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr7_-_3884424 | 2.67 |
ENSDART00000145641
|
si:dkey-88n24.3
|
si:dkey-88n24.3 |
| chr20_+_28861435 | 2.66 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr5_-_34993242 | 2.66 |
ENSDART00000134516
ENSDART00000051295 |
btf3
|
basic transcription factor 3 |
| chr5_-_42272517 | 2.59 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr19_+_12237945 | 2.59 |
ENSDART00000190034
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr20_+_18994059 | 2.57 |
ENSDART00000152380
|
tdh
|
L-threonine dehydrogenase |
| chr13_+_25412307 | 2.55 |
ENSDART00000175339
|
calhm1
|
calcium homeostasis modulator 1 |
| chr1_+_58137020 | 2.55 |
ENSDART00000180887
|
si:ch211-15j1.4
|
si:ch211-15j1.4 |
| chr6_+_56147812 | 2.54 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr16_-_2870522 | 2.52 |
ENSDART00000148543
|
cdcp1a
|
CUB domain containing protein 1a |
| chr5_+_45322734 | 2.50 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr6_-_54111928 | 2.49 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr14_-_6727717 | 2.48 |
ENSDART00000166979
|
si:dkeyp-44a8.4
|
si:dkeyp-44a8.4 |
| chr7_-_3792765 | 2.47 |
ENSDART00000060333
|
si:dkey-28d5.12
|
si:dkey-28d5.12 |
| chr7_+_4865367 | 2.45 |
ENSDART00000139120
|
si:dkey-28d5.8
|
si:dkey-28d5.8 |
| chr1_-_51720633 | 2.41 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr7_-_57637779 | 2.40 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr19_+_7864767 | 2.34 |
ENSDART00000137540
ENSDART00000151642 |
si:dkeyp-85e10.3
|
si:dkeyp-85e10.3 |
| chr8_+_30747940 | 2.33 |
ENSDART00000098975
|
p2rx4b
|
purinergic receptor P2X, ligand-gated ion channel, 4b |
| chr10_-_25817854 | 2.32 |
ENSDART00000147677
|
postna
|
periostin, osteoblast specific factor a |
| chr24_+_23791758 | 2.32 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr9_+_33423170 | 2.30 |
ENSDART00000141542
|
si:dkey-216e24.9
|
si:dkey-216e24.9 |
| chr1_-_51038885 | 2.30 |
ENSDART00000035150
|
spast
|
spastin |
| chr20_+_25626479 | 2.27 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr18_+_49969568 | 2.24 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr16_+_38167883 | 2.24 |
ENSDART00000111796
|
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr14_-_4145594 | 2.24 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr15_-_25556074 | 2.23 |
ENSDART00000124677
|
mmp20a
|
matrix metallopeptidase 20a (enamelysin) |
| chr23_+_11347313 | 2.20 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr15_+_1148074 | 2.17 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr12_+_48220584 | 2.15 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr21_-_11655010 | 2.11 |
ENSDART00000144370
ENSDART00000139814 ENSDART00000139289 |
cast
|
calpastatin |
| chr7_+_4682458 | 2.07 |
ENSDART00000139271
|
si:ch211-225k7.2
|
si:ch211-225k7.2 |
| chr7_-_3792972 | 2.07 |
ENSDART00000146304
|
si:dkey-28d5.12
|
si:dkey-28d5.12 |
| chr18_+_30998472 | 2.05 |
ENSDART00000154993
ENSDART00000099333 |
cd151l
|
CD151 antigen, like |
| chr20_-_38746889 | 2.01 |
ENSDART00000140275
|
trim54
|
tripartite motif containing 54 |
| chr23_-_32092443 | 2.01 |
ENSDART00000133688
|
letmd1
|
LETM1 domain containing 1 |
| chr3_-_32337653 | 2.00 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr2_+_20926202 | 2.00 |
ENSDART00000178880
|
pla2g4aa
|
phospholipase A2, group IVAa (cytosolic, calcium-dependent) |
| chr20_-_25626693 | 2.00 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr14_-_38929885 | 1.99 |
ENSDART00000148737
|
btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr22_-_20376488 | 1.98 |
ENSDART00000140187
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr7_+_34305903 | 1.96 |
ENSDART00000173575
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr7_+_4911590 | 1.95 |
ENSDART00000038628
|
si:dkey-28d5.11
|
si:dkey-28d5.11 |
| chr9_-_49493305 | 1.94 |
ENSDART00000148707
ENSDART00000148561 |
xirp2b
|
xin actin binding repeat containing 2b |
| chr7_+_44593756 | 1.94 |
ENSDART00000125365
|
si:ch211-189a15.5
|
si:ch211-189a15.5 |
| chr13_+_45582391 | 1.92 |
ENSDART00000058093
|
ldlrap1b
|
low density lipoprotein receptor adaptor protein 1b |
| chr4_-_9592402 | 1.92 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr7_-_3748702 | 1.90 |
ENSDART00000134839
|
si:ch211-282j17.10
|
si:ch211-282j17.10 |
| chr7_-_3649414 | 1.88 |
ENSDART00000064279
|
si:ch211-282j17.2
|
si:ch211-282j17.2 |
| chr1_+_51039558 | 1.88 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr8_+_28629741 | 1.86 |
ENSDART00000163915
|
si:dkey-109a10.2
|
si:dkey-109a10.2 |
| chr22_-_14161309 | 1.85 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr4_+_31259 | 1.84 |
ENSDART00000166826
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr20_+_28861629 | 1.84 |
ENSDART00000187274
ENSDART00000047826 |
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr16_+_19637384 | 1.83 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr13_-_30996072 | 1.79 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr14_+_35428152 | 1.78 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr16_-_46587938 | 1.78 |
ENSDART00000181433
|
BX323793.1
|
|
| chr16_+_7991274 | 1.78 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr4_+_77957611 | 1.78 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr25_-_18739924 | 1.77 |
ENSDART00000156328
|
si:dkeyp-93a5.2
|
si:dkeyp-93a5.2 |
| chr8_+_20994433 | 1.77 |
ENSDART00000141736
|
si:dkeyp-82a1.8
|
si:dkeyp-82a1.8 |
| chr3_+_41558682 | 1.75 |
ENSDART00000157023
|
card11
|
caspase recruitment domain family, member 11 |
| chr8_+_7315625 | 1.75 |
ENSDART00000135655
ENSDART00000181048 |
selenoh
|
selenoprotein H |
| chr7_-_3849926 | 1.74 |
ENSDART00000143064
|
si:dkey-88n24.7
|
si:dkey-88n24.7 |
| chr25_-_3830272 | 1.73 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr7_-_3630019 | 1.73 |
ENSDART00000138164
ENSDART00000064277 |
si:dkey-192d15.3
|
si:dkey-192d15.3 |
| chr7_+_4624926 | 1.72 |
ENSDART00000137387
|
si:dkey-83f18.6
|
si:dkey-83f18.6 |
| chr12_+_37401331 | 1.71 |
ENSDART00000125040
|
si:ch211-152f22.4
|
si:ch211-152f22.4 |
| chr7_-_3850090 | 1.68 |
ENSDART00000113403
ENSDART00000137683 ENSDART00000049926 |
si:dkey-28d5.12
si:dkey-88n24.7
|
si:dkey-28d5.12 si:dkey-88n24.7 |
| chr12_+_4900981 | 1.68 |
ENSDART00000171507
|
cd79b
|
CD79b molecule, immunoglobulin-associated beta |
| chr21_+_45685757 | 1.66 |
ENSDART00000160530
|
sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr7_+_4987530 | 1.64 |
ENSDART00000132983
ENSDART00000137067 |
si:dkey-81n2.1
|
si:dkey-81n2.1 |
| chr18_-_3520358 | 1.64 |
ENSDART00000181412
|
capn5a
|
calpain 5a |
| chr7_-_3723144 | 1.64 |
ENSDART00000138945
|
si:ch211-282j17.8
|
si:ch211-282j17.8 |
| chr19_+_19759577 | 1.62 |
ENSDART00000169480
|
hoxa5a
|
homeobox A5a |
| chr13_-_2112450 | 1.61 |
ENSDART00000189343
|
fam83b
|
family with sequence similarity 83, member B |
| chr13_-_33170733 | 1.61 |
ENSDART00000057382
|
fbln5
|
fibulin 5 |
| chr10_-_1276046 | 1.60 |
ENSDART00000169779
|
pdlim5b
|
PDZ and LIM domain 5b |
| chr7_-_4315859 | 1.59 |
ENSDART00000172762
|
si:ch211-63p21.8
|
si:ch211-63p21.8 |
| chr19_+_3653976 | 1.58 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr17_-_31579715 | 1.58 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr12_-_4176329 | 1.57 |
ENSDART00000092746
|
si:dkey-32n7.7
|
si:dkey-32n7.7 |
| chr3_+_23731109 | 1.55 |
ENSDART00000131410
|
hoxb3a
|
homeobox B3a |
| chr1_+_57176256 | 1.54 |
ENSDART00000152741
|
si:dkey-27j5.6
|
si:dkey-27j5.6 |
| chr21_+_31253048 | 1.52 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr15_-_4580763 | 1.52 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr7_-_6553460 | 1.52 |
ENSDART00000172903
|
si:ch1073-220m6.1
|
si:ch1073-220m6.1 |
| chr9_-_40073255 | 1.50 |
ENSDART00000189139
|
IKZF2
|
si:zfos-1425h8.1 |
| chr7_+_4854690 | 1.49 |
ENSDART00000131645
|
si:dkey-28d5.7
|
si:dkey-28d5.7 |
| chr8_-_2616326 | 1.47 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr3_-_15119856 | 1.44 |
ENSDART00000138328
|
xpo6
|
exportin 6 |
| chr12_+_46745239 | 1.43 |
ENSDART00000057179
|
plaub
|
plasminogen activator, urokinase b |
| chr19_+_19756425 | 1.42 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr21_+_26532789 | 1.40 |
ENSDART00000144627
|
vegfbb
|
vascular endothelial growth factor Bb |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 2.4 | 14.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 2.2 | 6.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.3 | 5.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 1.1 | 7.7 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 1.0 | 8.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 1.0 | 4.8 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.8 | 2.3 | GO:0034214 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.7 | 5.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.7 | 5.4 | GO:0031179 | peptide modification(GO:0031179) |
| 0.6 | 3.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.6 | 9.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.6 | 4.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.5 | 3.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.5 | 3.8 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.5 | 4.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.4 | 2.7 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.4 | 16.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.4 | 2.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 1.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.4 | 5.7 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.3 | 1.0 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.3 | 4.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 1.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 4.6 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.3 | 5.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 2.0 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.3 | 1.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.3 | 0.9 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.3 | 0.8 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.3 | 2.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 1.9 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.3 | 8.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 0.8 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.3 | 4.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 4.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 3.0 | GO:0031128 | developmental induction(GO:0031128) Spemann organizer formation(GO:0060061) |
| 0.2 | 3.8 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 1.5 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 2.4 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 3.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 3.0 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 3.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 4.4 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.2 | 2.6 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.2 | 0.8 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 2.6 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 3.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 2.3 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 2.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 3.4 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.2 | 2.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.2 | 2.8 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.2 | 11.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.2 | 3.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.4 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 6.1 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.1 | 0.8 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 3.5 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 1.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 4.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.3 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.1 | 2.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 5.2 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 0.4 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 2.7 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 2.4 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.1 | 2.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.6 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 2.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 1.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 1.2 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 2.2 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 1.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.5 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 1.2 | GO:0036302 | atrioventricular valve morphogenesis(GO:0003181) atrioventricular canal development(GO:0036302) |
| 0.1 | 1.0 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.6 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.8 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.3 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 1.4 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.2 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 4.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.3 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 1.1 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.9 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 3.2 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 3.5 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 1.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.8 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.0 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.8 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.9 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 1.7 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.6 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 1.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 2.1 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.9 | 6.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.6 | 2.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 11.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 2.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.4 | 1.7 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.3 | 4.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 1.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 0.8 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 12.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 6.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 6.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 4.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 11.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 4.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 5.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 4.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 7.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 2.0 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 2.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 4.2 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 5.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 13.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 3.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 4.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.8 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 2.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 2.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.9 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 15.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 1.3 | 12.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 1.2 | 4.8 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 1.0 | 4.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.9 | 4.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.8 | 3.8 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.7 | 4.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.7 | 5.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.7 | 2.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 2.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.6 | 5.4 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.6 | 2.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.5 | 2.0 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.5 | 5.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 2.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.4 | 1.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.4 | 5.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 1.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.4 | 13.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 2.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 2.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 1.4 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.3 | 3.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 3.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 1.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.2 | 2.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 2.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 1.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 4.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 1.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 11.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 4.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.8 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 4.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.6 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 2.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 1.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.0 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.3 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 9.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 5.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 1.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 7.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 5.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 5.6 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 7.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 15.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 5.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 2.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 1.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 5.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.3 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 5.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 6.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 11.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 4.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 4.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 0.8 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 2.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 2.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 4.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 4.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 4.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 3.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 6.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.0 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 2.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 1.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 3.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 3.1 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.2 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 1.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 2.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 4.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 3.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |