Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
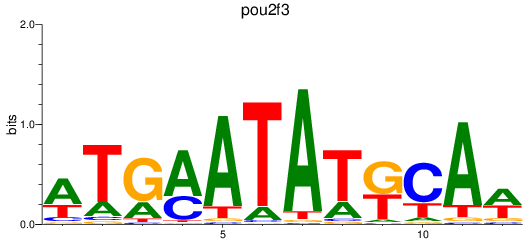
Results for pou2f3
Z-value: 0.95
Transcription factors associated with pou2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f3
|
ENSDARG00000052387 | POU class 2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f3 | dr11_v1_chr5_+_58550795_58550795 | 0.35 | 5.4e-04 | Click! |
Activity profile of pou2f3 motif
Sorted Z-values of pou2f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_46357080 | 12.13 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr18_-_20869175 | 11.60 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr13_+_22476742 | 10.83 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr15_+_46356879 | 9.98 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr20_+_572037 | 7.51 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr6_+_6924637 | 6.60 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr25_+_35019693 | 5.01 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr5_-_25583125 | 4.96 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr10_+_26597990 | 4.95 |
ENSDART00000079187
|
fhl1b
|
four and a half LIM domains 1b |
| chr5_-_25582721 | 4.75 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr24_+_35387517 | 4.51 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr11_+_30647545 | 4.42 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr3_+_39566999 | 4.42 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr10_+_39084354 | 4.36 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr2_+_15048410 | 4.32 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr22_+_5120033 | 4.32 |
ENSDART00000169200
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr22_-_26595027 | 4.22 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr25_+_35189555 | 4.19 |
ENSDART00000044453
|
ano5a
|
anoctamin 5a |
| chr4_+_7391110 | 4.14 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr21_+_43328685 | 4.03 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr9_-_30165621 | 3.98 |
ENSDART00000089543
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr19_-_7450796 | 3.86 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr9_-_34396264 | 3.83 |
ENSDART00000045754
|
grtp1b
|
growth hormone regulated TBC protein 1b |
| chr17_-_37395460 | 3.83 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr13_-_2189761 | 3.77 |
ENSDART00000166255
|
mlip
|
muscular LMNA-interacting protein |
| chr15_-_43284021 | 3.67 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr25_+_31227747 | 3.62 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr3_-_50865079 | 3.51 |
ENSDART00000164295
|
pmp22a
|
peripheral myelin protein 22a |
| chr10_+_5268054 | 3.39 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr2_-_16159491 | 3.39 |
ENSDART00000110059
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr10_-_36793412 | 3.26 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr3_+_49021079 | 3.25 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr25_+_3058924 | 3.20 |
ENSDART00000029580
|
fth1b
|
ferritin, heavy polypeptide 1b |
| chr22_+_28446557 | 3.19 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr13_-_44423 | 3.16 |
ENSDART00000093247
|
gtf2a1l
|
general transcription factor IIA, 1-like |
| chr18_-_12327426 | 3.02 |
ENSDART00000136992
ENSDART00000114024 |
fam107b
|
family with sequence similarity 107, member B |
| chr13_-_2215213 | 3.02 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr20_+_23625387 | 2.97 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr3_+_45687266 | 2.88 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr19_-_25114701 | 2.86 |
ENSDART00000149035
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr25_-_23526058 | 2.83 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr9_-_6380653 | 2.81 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr25_+_31277415 | 2.81 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr20_+_40150612 | 2.80 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr14_+_48862987 | 2.73 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr10_+_2582254 | 2.67 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr14_-_17622080 | 2.66 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr10_+_35439952 | 2.62 |
ENSDART00000006284
|
hhla2a.2
|
HERV-H LTR-associating 2a, tandem duplicate 2 |
| chr2_-_16159203 | 2.53 |
ENSDART00000153480
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr8_+_22516728 | 2.48 |
ENSDART00000146013
|
si:ch211-261n11.3
|
si:ch211-261n11.3 |
| chr5_+_27404946 | 2.47 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr7_+_20017211 | 2.46 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr17_+_38573471 | 2.45 |
ENSDART00000040627
|
sptb
|
spectrin, beta, erythrocytic |
| chr6_-_20875111 | 2.45 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr10_+_9595575 | 2.44 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr8_-_39754621 | 2.42 |
ENSDART00000149243
|
si:ch211-170d8.8
|
si:ch211-170d8.8 |
| chr19_-_5345930 | 2.38 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr22_+_38049130 | 2.35 |
ENSDART00000097533
|
wwtr1
|
WW domain containing transcription regulator 1 |
| chr23_+_11285662 | 2.34 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr15_+_46344655 | 2.22 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr2_+_36015049 | 2.21 |
ENSDART00000158276
|
lamc2
|
laminin, gamma 2 |
| chr18_-_15551360 | 2.20 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr2_+_32796873 | 2.19 |
ENSDART00000077511
|
ccr9a
|
chemokine (C-C motif) receptor 9a |
| chr19_-_22387141 | 2.18 |
ENSDART00000151234
|
eppk1
|
epiplakin 1 |
| chr4_+_4509996 | 2.15 |
ENSDART00000028694
|
gnsa
|
glucosamine (N-acetyl)-6-sulfatase a |
| chr2_+_6243144 | 2.12 |
ENSDART00000058258
|
gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr2_+_20605186 | 2.11 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr9_+_3388099 | 2.10 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr14_+_3495542 | 2.07 |
ENSDART00000168934
|
gstp2
|
glutathione S-transferase pi 2 |
| chr23_-_33709964 | 2.07 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr1_-_38815361 | 2.06 |
ENSDART00000148790
ENSDART00000148572 ENSDART00000149080 |
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr22_+_14051894 | 2.05 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr11_-_36341028 | 2.03 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr17_-_6451801 | 1.99 |
ENSDART00000064700
|
fuca2
|
alpha-L-fucosidase 2 |
| chr17_-_29213710 | 1.99 |
ENSDART00000076481
|
ehd4
|
EH-domain containing 4 |
| chr7_-_2116512 | 1.98 |
ENSDART00000098148
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr23_+_22200467 | 1.97 |
ENSDART00000025414
|
slc2a1a
|
solute carrier family 2 (facilitated glucose transporter), member 1a |
| chr7_-_18168493 | 1.95 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr2_+_20604775 | 1.95 |
ENSDART00000131501
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr17_-_25382367 | 1.94 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr20_+_26967154 | 1.94 |
ENSDART00000153294
ENSDART00000132434 ENSDART00000143047 |
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr7_+_38897836 | 1.93 |
ENSDART00000024330
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr1_+_21563311 | 1.88 |
ENSDART00000147076
ENSDART00000006147 |
primpol
|
primase and polymerase (DNA-directed) |
| chr10_+_18911152 | 1.88 |
ENSDART00000030205
|
bnip3lb
|
BCL2 interacting protein 3 like b |
| chr8_+_52479026 | 1.87 |
ENSDART00000162885
|
FO704661.1
|
Danio rerio gamma-glutamyl hydrolase (LOC553228), mRNA. |
| chr20_-_45812144 | 1.87 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr13_-_30645965 | 1.86 |
ENSDART00000109307
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr19_-_25772980 | 1.84 |
ENSDART00000052393
|
pard6gb
|
par-6 family cell polarity regulator gamma b |
| chr8_+_36268747 | 1.80 |
ENSDART00000183154
|
CU929676.1
|
|
| chr15_-_31514818 | 1.80 |
ENSDART00000153978
|
hmgb1b
|
high mobility group box 1b |
| chr23_+_33907899 | 1.79 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr6_-_53048291 | 1.78 |
ENSDART00000103267
|
fam212ab
|
family with sequence similarity 212, member Ab |
| chr25_+_34915576 | 1.75 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr14_+_15231097 | 1.74 |
ENSDART00000172430
|
si:dkey-203a12.3
|
si:dkey-203a12.3 |
| chr21_+_34088377 | 1.73 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr3_-_5067585 | 1.73 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr7_-_48667056 | 1.72 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr8_-_23573084 | 1.72 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr11_-_36341189 | 1.69 |
ENSDART00000159752
|
sort1a
|
sortilin 1a |
| chr19_+_1688727 | 1.69 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr6_-_54796072 | 1.68 |
ENSDART00000164445
|
tnnt2b
|
troponin T type 2b (cardiac) |
| chr1_+_51496862 | 1.67 |
ENSDART00000150433
|
meis1a
|
Meis homeobox 1 a |
| chr8_-_19977088 | 1.67 |
ENSDART00000139647
|
lpxn
|
leupaxin |
| chr10_+_26667475 | 1.66 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr3_+_48473346 | 1.65 |
ENSDART00000166294
|
metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr19_-_47523793 | 1.64 |
ENSDART00000124234
|
grem1a
|
gremlin 1a, DAN family BMP antagonist |
| chr3_-_39305291 | 1.64 |
ENSDART00000102674
|
plcd3a
|
phospholipase C, delta 3a |
| chr20_-_35750810 | 1.63 |
ENSDART00000153072
|
adgrf8
|
adhesion G protein-coupled receptor F8 |
| chr2_-_3419890 | 1.62 |
ENSDART00000055618
|
iba57
|
IBA57, iron-sulfur cluster assembly |
| chr16_-_42523744 | 1.62 |
ENSDART00000017185
|
tbx20
|
T-box 20 |
| chr16_-_26140768 | 1.59 |
ENSDART00000143960
|
cd79a
|
CD79a molecule, immunoglobulin-associated alpha |
| chr21_+_34088110 | 1.57 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr25_-_20666754 | 1.56 |
ENSDART00000158418
|
csk
|
C-terminal Src kinase |
| chr19_-_340641 | 1.55 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr10_-_22918214 | 1.54 |
ENSDART00000163908
|
rnasekb
|
ribonuclease, RNase K b |
| chr1_-_44161417 | 1.52 |
ENSDART00000083213
|
slc43a3a
|
solute carrier family 43, member 3a |
| chr4_+_12292274 | 1.50 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr19_-_205104 | 1.50 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr18_-_14677936 | 1.48 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr3_-_59981476 | 1.48 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr18_+_27318589 | 1.47 |
ENSDART00000037813
|
cd81b
|
CD81 molecule b |
| chr2_-_23778180 | 1.46 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr9_-_8979468 | 1.45 |
ENSDART00000134646
|
ing1
|
inhibitor of growth family, member 1 |
| chr6_+_36381709 | 1.45 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr23_-_35066816 | 1.44 |
ENSDART00000168731
ENSDART00000163731 |
BX294434.1
|
|
| chr17_+_24109012 | 1.43 |
ENSDART00000156251
|
ehbp1
|
EH domain binding protein 1 |
| chr10_-_31015535 | 1.43 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr22_-_19552796 | 1.43 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr4_-_12388535 | 1.41 |
ENSDART00000017180
|
rergla
|
RERG/RAS-like a |
| chr17_-_50311009 | 1.41 |
ENSDART00000153653
|
si:ch73-50f9.4
|
si:ch73-50f9.4 |
| chr12_+_17504559 | 1.41 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr1_-_41287914 | 1.40 |
ENSDART00000022546
ENSDART00000126853 ENSDART00000125988 |
rgs12b
|
regulator of G protein signaling 12b |
| chr16_-_35952789 | 1.39 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr9_-_1951144 | 1.37 |
ENSDART00000082355
|
hoxd4a
|
homeobox D4a |
| chr1_-_51038885 | 1.36 |
ENSDART00000035150
|
spast
|
spastin |
| chr25_+_34915762 | 1.36 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr5_-_23909934 | 1.35 |
ENSDART00000142516
|
si:ch211-135f11.1
|
si:ch211-135f11.1 |
| chr12_-_48188928 | 1.34 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr5_-_32274383 | 1.34 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr11_+_37275448 | 1.33 |
ENSDART00000161423
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr3_-_45361573 | 1.33 |
ENSDART00000154796
|
il21r.1
|
interleukin 21 receptor, tandem duplicate 1 |
| chr5_+_51848756 | 1.32 |
ENSDART00000087467
ENSDART00000184466 |
cmya5
|
cardiomyopathy associated 5 |
| chr2_+_55365727 | 1.32 |
ENSDART00000162943
|
FP245456.1
|
|
| chr7_-_44704910 | 1.31 |
ENSDART00000037850
|
dync1li2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr20_+_29691118 | 1.30 |
ENSDART00000164121
|
mboat2b
|
membrane bound O-acyltransferase domain containing 2b |
| chr18_+_30567945 | 1.28 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr1_-_51720633 | 1.25 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr2_-_48171112 | 1.23 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr6_-_30932078 | 1.22 |
ENSDART00000028612
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr11_+_1608348 | 1.22 |
ENSDART00000162438
|
si:dkey-40c23.3
|
si:dkey-40c23.3 |
| chr6_-_35032792 | 1.22 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr11_+_10984293 | 1.21 |
ENSDART00000065933
|
itgb6
|
integrin, beta 6 |
| chr5_-_24127310 | 1.20 |
ENSDART00000182700
ENSDART00000154313 |
capga
|
capping protein (actin filament), gelsolin-like a |
| chr23_-_33775145 | 1.18 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr7_+_30725473 | 1.18 |
ENSDART00000085716
|
mtmr10
|
myotubularin related protein 10 |
| chr10_+_13000370 | 1.18 |
ENSDART00000180545
|
lpar1
|
lysophosphatidic acid receptor 1 |
| chr10_+_39893439 | 1.17 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr1_-_46989650 | 1.17 |
ENSDART00000129072
ENSDART00000150507 |
si:dkey-22n8.3
|
si:dkey-22n8.3 |
| chr24_-_28333029 | 1.16 |
ENSDART00000149015
ENSDART00000129174 |
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr10_+_26871751 | 1.16 |
ENSDART00000089205
|
ehbp1l1b
|
EH domain binding protein 1-like 1b |
| chr25_-_18140305 | 1.16 |
ENSDART00000180222
|
kitlga
|
kit ligand a |
| chr19_+_43579786 | 1.15 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr11_-_43200994 | 1.14 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr22_+_9091913 | 1.13 |
ENSDART00000186598
|
BX248395.1
|
|
| chr20_-_31497300 | 1.13 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr1_+_51039558 | 1.13 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr9_+_21281652 | 1.11 |
ENSDART00000113352
|
lats2
|
large tumor suppressor kinase 2 |
| chr7_+_38898208 | 1.10 |
ENSDART00000172251
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr4_-_67980261 | 1.09 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr5_-_54481692 | 1.08 |
ENSDART00000165719
|
fbxw5
|
F-box and WD repeat domain containing 5 |
| chr19_-_30811161 | 1.08 |
ENSDART00000103524
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr13_-_34781984 | 1.07 |
ENSDART00000172138
|
ism1
|
isthmin 1 |
| chr12_-_11593436 | 1.06 |
ENSDART00000138954
|
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr11_-_42918971 | 1.06 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr8_+_49433663 | 1.06 |
ENSDART00000140481
ENSDART00000180714 |
nfu1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr18_-_21950751 | 1.05 |
ENSDART00000043452
|
tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr4_+_25950372 | 1.04 |
ENSDART00000125767
|
metap2a
|
methionyl aminopeptidase 2a |
| chr19_-_5103313 | 1.03 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr14_-_17588345 | 1.02 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr24_-_25428176 | 1.02 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr1_+_1896737 | 1.02 |
ENSDART00000152442
|
si:ch211-132g1.6
|
si:ch211-132g1.6 |
| chr20_-_54198130 | 1.01 |
ENSDART00000160409
|
arf6a
|
ADP-ribosylation factor 6a |
| chr20_-_46817223 | 1.01 |
ENSDART00000100336
|
esrrgb
|
estrogen-related receptor gamma b |
| chr3_+_14543953 | 1.01 |
ENSDART00000161710
|
epor
|
erythropoietin receptor |
| chr12_+_2677303 | 1.01 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr18_+_2222447 | 1.00 |
ENSDART00000185927
|
pigbos1
|
PIGB opposite strand 1 |
| chr21_-_37733287 | 0.99 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr6_-_24053404 | 0.99 |
ENSDART00000168511
|
si:dkey-44g17.6
|
si:dkey-44g17.6 |
| chr16_-_38333976 | 0.98 |
ENSDART00000031895
|
cdc42se1
|
CDC42 small effector 1 |
| chr25_+_32496723 | 0.98 |
ENSDART00000087978
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr12_-_10409961 | 0.96 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr7_-_58098814 | 0.96 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr14_-_33894915 | 0.95 |
ENSDART00000143290
|
urp1
|
urotensin-related peptide 1 |
| chr25_+_19238175 | 0.94 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr25_-_20666328 | 0.94 |
ENSDART00000098076
|
csk
|
C-terminal Src kinase |
| chr17_+_4400738 | 0.94 |
ENSDART00000170204
|
si:zfos-364h11.1
|
si:zfos-364h11.1 |
| chr25_+_16601839 | 0.93 |
ENSDART00000008986
|
atp6v1e1a
|
ATPase H+ transporting V1 subunit E1a |
| chr15_-_23482088 | 0.93 |
ENSDART00000185823
ENSDART00000185523 |
nlrx1
|
NLR family member X1 |
| chr17_-_30975978 | 0.93 |
ENSDART00000051697
|
evla
|
Enah/Vasp-like a |
| chr21_-_25295087 | 0.92 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.7 | GO:0042088 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.3 | 3.8 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.9 | 7.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.9 | 2.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.8 | 3.9 | GO:0090200 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.6 | 1.9 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.6 | 2.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.6 | 5.0 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.5 | 2.5 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.5 | 2.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.5 | 3.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.5 | 3.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 1.4 | GO:0051230 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.4 | 1.3 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.4 | 1.6 | GO:0055021 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.4 | 2.0 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.4 | 1.5 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 1.9 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.4 | 2.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 1.6 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.3 | 2.2 | GO:0045109 | intermediate filament organization(GO:0045109) intermediate filament bundle assembly(GO:0045110) |
| 0.3 | 0.9 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.3 | 2.0 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.3 | 2.0 | GO:0046323 | glucose import(GO:0046323) |
| 0.3 | 1.9 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.3 | 1.3 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.3 | 1.0 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.3 | 1.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 1.7 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 2.6 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.2 | 5.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 0.7 | GO:2000405 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.2 | 2.2 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 0.6 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 1.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 10.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 5.3 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.2 | 0.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 2.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 0.4 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.2 | 3.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 1.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 1.9 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.2 | 0.5 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 0.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.9 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 2.1 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.2 | 2.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 1.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.0 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 1.0 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 2.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.6 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 0.4 | GO:0032816 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.1 | 1.2 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.3 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.1 | 1.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 1.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.7 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 1.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.3 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 1.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.4 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.5 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 2.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 4.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.6 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.1 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 4.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.5 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 1.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 3.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.9 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.4 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 0.7 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 2.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.2 | GO:0046833 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 6.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 9.2 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.1 | 1.7 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.1 | 1.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 3.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 1.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 1.1 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.1 | 1.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.6 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.6 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 0.3 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.3 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.5 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 4.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.5 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 2.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 2.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 1.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.4 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.3 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.0 | 2.4 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 3.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 1.2 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.1 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 1.9 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 3.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.4 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 1.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 4.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.5 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 3.1 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 5.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 2.1 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 1.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 2.5 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 0.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 1.9 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.7 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 2.2 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.7 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 1.4 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.3 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.8 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 1.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.7 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.1 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.0 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.7 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:1902804 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 1.1 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.7 | 3.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.6 | 9.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.4 | 1.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 1.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.3 | 1.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 14.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.3 | 12.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 3.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 0.7 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 6.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 13.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 5.5 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 3.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 3.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 1.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 2.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 4.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.7 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 3.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 1.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.9 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.2 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.2 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 1.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 2.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 3.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.5 | GO:0098844 | dendritic spine head(GO:0044327) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 3.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 8.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.5 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 1.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 4.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.8 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 3.7 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:0005521 | lamin binding(GO:0005521) |
| 1.6 | 9.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.7 | 2.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.7 | 2.0 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.6 | 4.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.6 | 4.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.5 | 10.8 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.5 | 7.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.4 | 1.5 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 2.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 2.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 1.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.3 | 3.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 3.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 1.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 3.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 0.9 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 5.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 1.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 0.6 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.2 | 1.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 1.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 0.9 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 1.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 3.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 1.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 1.7 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 2.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 3.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.4 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 1.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.9 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 2.0 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 2.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.9 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 2.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 4.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.9 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 2.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 0.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 1.2 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.1 | 0.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 3.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 4.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.0 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 3.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 4.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.8 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 1.2 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.8 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 1.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 3.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 4.6 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 3.3 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 1.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.9 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 5.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 6.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 2.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 5.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 2.5 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 1.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.3 | 3.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 3.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |