Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
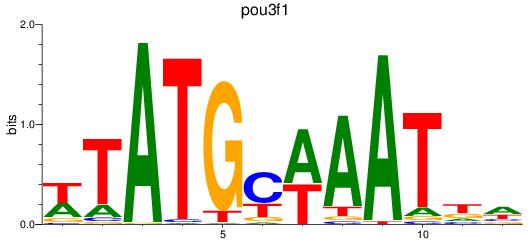
Results for pou3f1
Z-value: 0.88
Transcription factors associated with pou3f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou3f1
|
ENSDARG00000009823 | POU class 3 homeobox 1 |
|
pou3f1
|
ENSDARG00000112302 | POU class 3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou3f1 | dr11_v1_chr16_+_33593116_33593116 | 0.58 | 6.4e-10 | Click! |
Activity profile of pou3f1 motif
Sorted Z-values of pou3f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_30563115 | 15.13 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr19_-_30562648 | 14.19 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr22_+_20195280 | 12.83 |
ENSDART00000088603
ENSDART00000135692 |
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr10_+_22003750 | 11.32 |
ENSDART00000109420
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr22_+_5106751 | 11.21 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr16_-_12173554 | 10.80 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr6_+_4539953 | 10.57 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr23_+_28731379 | 10.28 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr7_+_40228422 | 9.48 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr16_-_43971258 | 9.29 |
ENSDART00000141941
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr9_-_18877597 | 9.13 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr7_-_38633867 | 9.11 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr17_-_15528597 | 8.59 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr6_-_13187168 | 8.53 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr21_-_27185915 | 7.78 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr19_-_28367413 | 7.76 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr7_-_24699985 | 7.62 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr18_-_36135799 | 7.55 |
ENSDART00000059344
|
b3gat1a
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) a |
| chr5_-_24000211 | 7.55 |
ENSDART00000188865
|
map7d2a
|
MAP7 domain containing 2a |
| chr23_+_44307996 | 7.29 |
ENSDART00000042430
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr2_-_34555945 | 7.18 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr3_+_33345348 | 7.08 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr18_+_38749547 | 7.04 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr2_+_24203229 | 7.02 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr23_-_31372639 | 6.84 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr13_+_33117528 | 6.82 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr6_-_13188667 | 6.78 |
ENSDART00000191654
|
adam23a
|
ADAM metallopeptidase domain 23a |
| chr16_+_39159752 | 6.74 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr23_+_36771593 | 6.63 |
ENSDART00000078240
|
march9
|
membrane-associated ring finger (C3HC4) 9 |
| chr6_-_13308813 | 6.42 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr23_+_28582865 | 5.98 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr21_+_41743493 | 5.83 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr16_+_43152727 | 5.70 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr1_+_1599979 | 5.70 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr20_+_27298783 | 5.66 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr4_-_8903240 | 5.50 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr18_+_7286788 | 5.43 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr5_-_16996482 | 5.35 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr21_+_26390549 | 5.34 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr16_-_12173399 | 5.15 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr25_-_15045338 | 4.96 |
ENSDART00000161165
ENSDART00000165774 ENSDART00000172538 |
pax6a
|
paired box 6a |
| chr2_+_2563192 | 4.81 |
ENSDART00000055713
|
CABZ01033000.1
|
|
| chr1_+_46194333 | 4.66 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr8_-_15398760 | 4.62 |
ENSDART00000162011
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr7_+_61050329 | 4.61 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr25_-_25736958 | 4.60 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr22_+_18389271 | 4.54 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr1_+_7679328 | 4.45 |
ENSDART00000163488
ENSDART00000190070 |
en1b
|
engrailed homeobox 1b |
| chr9_-_31915423 | 4.35 |
ENSDART00000060051
|
fgf14
|
fibroblast growth factor 14 |
| chr7_+_31074636 | 4.11 |
ENSDART00000173805
|
tjp1a
|
tight junction protein 1a |
| chr13_+_28705143 | 4.09 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr5_+_30624183 | 4.09 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr3_-_6519691 | 3.90 |
ENSDART00000165273
ENSDART00000179882 ENSDART00000172292 |
GGA3 (1 of many)
|
si:ch73-157i16.3 |
| chr14_-_40389699 | 3.80 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr12_+_28986308 | 3.71 |
ENSDART00000153178
|
si:rp71-1c10.8
|
si:rp71-1c10.8 |
| chr17_+_15882533 | 3.68 |
ENSDART00000164124
|
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr15_-_30815826 | 3.63 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr18_-_12957451 | 3.59 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr14_-_48939560 | 3.57 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr3_-_5964557 | 3.49 |
ENSDART00000184738
|
BX284638.1
|
|
| chr24_+_41931585 | 3.48 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr20_-_39188476 | 3.45 |
ENSDART00000152858
|
rcan2
|
regulator of calcineurin 2 |
| chr14_+_23874062 | 3.44 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr1_+_31725154 | 3.43 |
ENSDART00000112333
ENSDART00000189801 |
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr17_-_37052622 | 3.34 |
ENSDART00000186408
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr5_-_46896541 | 3.15 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr21_-_25573064 | 3.12 |
ENSDART00000134310
|
CR388166.1
|
|
| chr7_-_40738774 | 3.11 |
ENSDART00000084179
|
rnf32
|
ring finger protein 32 |
| chr6_+_45692026 | 2.98 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr16_+_45746549 | 2.97 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr11_-_35575791 | 2.97 |
ENSDART00000031441
ENSDART00000188513 ENSDART00000183609 |
sema3fb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fb |
| chr17_+_33719415 | 2.96 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr22_-_11729350 | 2.94 |
ENSDART00000105813
|
krt222
|
keratin 222 |
| chr19_-_19556778 | 2.93 |
ENSDART00000164060
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr24_-_37484123 | 2.93 |
ENSDART00000111623
|
cluap1
|
clusterin associated protein 1 |
| chr25_+_25737386 | 2.91 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr11_+_21586335 | 2.71 |
ENSDART00000091182
|
FAM72B
|
zgc:101564 |
| chr17_-_42213822 | 2.65 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr6_+_18418651 | 2.57 |
ENSDART00000171097
ENSDART00000167798 |
rab11fip4b
|
RAB11 family interacting protein 4 (class II) b |
| chr16_+_25074029 | 2.55 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr17_-_4245311 | 2.52 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr11_+_25634041 | 2.47 |
ENSDART00000033657
|
grm6b
|
glutamate receptor, metabotropic 6b |
| chr13_+_29778610 | 2.47 |
ENSDART00000132004
|
pax2a
|
paired box 2a |
| chr12_+_48480632 | 2.47 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr7_+_60396957 | 2.45 |
ENSDART00000121545
|
brms1
|
breast cancer metastasis suppressor 1 |
| chr7_+_25920792 | 2.41 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr9_-_18735256 | 2.41 |
ENSDART00000143165
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr15_-_5157572 | 2.41 |
ENSDART00000174192
|
or128-10
|
odorant receptor, family E, subfamily 128, member 10 |
| chr1_+_54013457 | 2.39 |
ENSDART00000012104
ENSDART00000126339 |
dla
|
deltaA |
| chr22_-_18491813 | 2.36 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr2_+_52847049 | 2.34 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr3_-_50998577 | 2.29 |
ENSDART00000157735
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr24_-_8730913 | 2.23 |
ENSDART00000187228
ENSDART00000082349 ENSDART00000186660 |
tfap2a
|
transcription factor AP-2 alpha |
| chr6_-_16406210 | 2.22 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr23_-_3409140 | 2.20 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr22_+_18786797 | 2.14 |
ENSDART00000141864
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr12_+_16087077 | 2.13 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr5_+_69733096 | 2.10 |
ENSDART00000169013
|
arl6ip4
|
ADP-ribosylation factor-like 6 interacting protein 4 |
| chr13_+_29773153 | 2.09 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr21_-_30254185 | 2.06 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr20_+_18225329 | 1.96 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr23_+_20518504 | 1.95 |
ENSDART00000114246
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr6_-_19381993 | 1.94 |
ENSDART00000164114
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr8_+_28469054 | 1.92 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr7_-_4036184 | 1.89 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr8_-_43574935 | 1.88 |
ENSDART00000051139
|
ncor2
|
nuclear receptor corepressor 2 |
| chr1_+_8984068 | 1.84 |
ENSDART00000122568
|
tlr2
|
toll-like receptor 2 |
| chr15_+_39096736 | 1.84 |
ENSDART00000129511
ENSDART00000014877 |
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr9_-_21507083 | 1.84 |
ENSDART00000137922
|
aff3
|
AF4/FMR2 family, member 3 |
| chr14_-_34700633 | 1.82 |
ENSDART00000150358
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr1_+_27825980 | 1.81 |
ENSDART00000160524
|
pspc1
|
paraspeckle component 1 |
| chr22_+_14051894 | 1.78 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr5_-_25406807 | 1.75 |
ENSDART00000089748
|
rorb
|
RAR-related orphan receptor B |
| chr23_+_2825940 | 1.74 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr19_-_26769867 | 1.68 |
ENSDART00000043776
ENSDART00000159489 ENSDART00000138675 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr6_+_41096058 | 1.62 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr10_+_39304422 | 1.56 |
ENSDART00000019267
|
kcnj1b
|
potassium inwardly-rectifying channel, subfamily J, member 1b |
| chr21_-_32467099 | 1.56 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr20_+_48782068 | 1.55 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr22_-_7400471 | 1.55 |
ENSDART00000161215
|
si:dkey-57c15.1
|
si:dkey-57c15.1 |
| chr19_-_26770083 | 1.54 |
ENSDART00000193811
ENSDART00000174455 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr1_-_20593778 | 1.52 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr2_-_56131312 | 1.50 |
ENSDART00000097755
|
jund
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr7_-_26497947 | 1.49 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr25_+_14694876 | 1.49 |
ENSDART00000050478
ENSDART00000188093 |
mpped2
|
metallophosphoesterase domain containing 2b |
| chr9_-_30502010 | 1.48 |
ENSDART00000149483
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr5_+_63785339 | 1.47 |
ENSDART00000050871
|
rgs3b
|
regulator of G protein signaling 3b |
| chr10_+_26972755 | 1.46 |
ENSDART00000042162
|
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr18_-_47103131 | 1.43 |
ENSDART00000188553
|
CABZ01039304.1
|
|
| chr15_+_34592215 | 1.43 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr11_-_41130239 | 1.41 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr5_+_63302660 | 1.40 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr24_-_40009446 | 1.38 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr8_+_31777633 | 1.38 |
ENSDART00000142519
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr7_+_69470442 | 1.35 |
ENSDART00000189593
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr25_-_26673570 | 1.35 |
ENSDART00000154917
|
ciartb
|
circadian associated repressor of transcription b |
| chr13_-_33822550 | 1.34 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr12_-_33579873 | 1.33 |
ENSDART00000184661
|
tdrkh
|
tudor and KH domain containing |
| chr2_-_39036604 | 1.32 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr25_+_5604512 | 1.31 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr17_-_25831569 | 1.31 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr10_-_5821584 | 1.29 |
ENSDART00000166388
|
il6st
|
interleukin 6 signal transducer |
| chr22_+_17784414 | 1.28 |
ENSDART00000188189
ENSDART00000145260 |
tm6sf2
|
transmembrane 6 superfamily member 2 |
| chr5_+_9147497 | 1.24 |
ENSDART00000003273
|
rchy1
|
ring finger and CHY zinc finger domain containing 1 |
| chr23_+_21473103 | 1.23 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr24_+_25467465 | 1.22 |
ENSDART00000189933
|
smpx
|
small muscle protein, X-linked |
| chr2_-_37684641 | 1.20 |
ENSDART00000012191
|
hiat1a
|
hippocampus abundant transcript 1a |
| chr3_+_30190419 | 1.17 |
ENSDART00000157320
|
akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr15_-_734203 | 1.17 |
ENSDART00000177867
ENSDART00000154918 |
si:dkey-7i4.21
BX004964.1
|
si:dkey-7i4.21 |
| chr1_-_16394814 | 1.16 |
ENSDART00000013024
|
fgf20a
|
fibroblast growth factor 20a |
| chr14_+_6991142 | 1.16 |
ENSDART00000157635
ENSDART00000059918 |
hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr17_+_43013171 | 1.13 |
ENSDART00000055541
|
gskip
|
gsk3b interacting protein |
| chr22_-_11954861 | 1.11 |
ENSDART00000131611
|
si:dkeyp-4h4.1
|
si:dkeyp-4h4.1 |
| chr10_+_26973063 | 1.11 |
ENSDART00000143162
ENSDART00000186210 |
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr10_+_42521943 | 1.11 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr1_-_17587552 | 1.08 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr9_+_12444494 | 1.03 |
ENSDART00000102430
|
tmem41aa
|
transmembrane protein 41aa |
| chr14_+_37545639 | 1.03 |
ENSDART00000173192
|
pcdh1b
|
protocadherin 1b |
| chr4_-_3353595 | 1.03 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_-_40411944 | 1.03 |
ENSDART00000144135
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr12_+_36092218 | 1.00 |
ENSDART00000168694
|
btbd17a
|
BTB (POZ) domain containing 17a |
| chr18_-_44359726 | 1.00 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr20_-_45060241 | 0.98 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr21_+_8427059 | 0.98 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr2_-_24317240 | 0.98 |
ENSDART00000078975
|
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr13_-_48756036 | 0.96 |
ENSDART00000035018
ENSDART00000132895 |
ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr23_-_6765653 | 0.96 |
ENSDART00000192310
|
FP102169.1
|
|
| chr10_+_44940693 | 0.94 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr9_-_25444091 | 0.94 |
ENSDART00000002428
|
acvr2aa
|
activin A receptor type 2Aa |
| chr22_-_10397600 | 0.93 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr13_-_24877577 | 0.89 |
ENSDART00000182705
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr11_+_25257022 | 0.88 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr20_+_20751425 | 0.88 |
ENSDART00000177048
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr22_+_413349 | 0.87 |
ENSDART00000082453
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr16_-_21620947 | 0.87 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_-_44876005 | 0.86 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr10_+_33895315 | 0.84 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr8_+_8238727 | 0.83 |
ENSDART00000144838
|
plxnb3
|
plexin B3 |
| chr16_+_25107344 | 0.83 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
| chr2_+_32835679 | 0.82 |
ENSDART00000132767
|
pxdc1a
|
PX domain containing 1a |
| chr7_+_20965880 | 0.81 |
ENSDART00000062003
|
efnb3b
|
ephrin-B3b |
| chr12_-_19151708 | 0.81 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr12_+_46960579 | 0.81 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr10_+_33744098 | 0.79 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr7_+_22792895 | 0.76 |
ENSDART00000184407
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr3_+_26223376 | 0.76 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr23_-_3408777 | 0.75 |
ENSDART00000193245
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr17_-_4245902 | 0.73 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr15_+_29727799 | 0.73 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr2_-_3616363 | 0.71 |
ENSDART00000144699
|
pter
|
phosphotriesterase related |
| chr4_+_12292274 | 0.70 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr2_-_51290607 | 0.69 |
ENSDART00000171693
|
si:ch211-215e19.3
|
si:ch211-215e19.3 |
| chr4_+_12013043 | 0.66 |
ENSDART00000130692
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr1_-_24918686 | 0.66 |
ENSDART00000148076
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr7_+_10701938 | 0.66 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr17_-_30473374 | 0.65 |
ENSDART00000155021
|
si:ch211-175f11.5
|
si:ch211-175f11.5 |
| chr10_-_26225548 | 0.64 |
ENSDART00000132019
ENSDART00000079194 |
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr22_-_32392252 | 0.64 |
ENSDART00000148681
ENSDART00000137945 |
pcbp4
|
poly(rC) binding protein 4 |
| chr16_+_38350704 | 0.63 |
ENSDART00000140761
|
elp6
|
elongator acetyltransferase complex subunit 6 |
| chr21_+_5888641 | 0.62 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou3f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 2.3 | 6.8 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 2.1 | 12.8 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 1.9 | 7.6 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 1.8 | 16.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 1.7 | 6.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 1.3 | 5.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.1 | 4.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 1.1 | 4.5 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 1.1 | 8.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 1.0 | 11.2 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.9 | 2.6 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.7 | 3.0 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.7 | 7.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.7 | 5.0 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.7 | 5.7 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.7 | 3.5 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.7 | 0.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.7 | 3.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.6 | 7.8 | GO:0042044 | fluid transport(GO:0042044) |
| 0.6 | 1.8 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.6 | 2.4 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.6 | 5.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.6 | 1.7 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.6 | 4.6 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.5 | 6.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.5 | 3.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.5 | 15.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.5 | 1.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.4 | 2.4 | GO:0055016 | hypochord development(GO:0055016) |
| 0.4 | 4.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.3 | 3.8 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 1.3 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.3 | 1.9 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.3 | 4.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 2.1 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.3 | 1.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.3 | 7.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.3 | 1.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.3 | 4.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.3 | 6.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 0.7 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.2 | 0.5 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.2 | 2.4 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.2 | 2.3 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 2.5 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 0.7 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.2 | 1.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 4.1 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.2 | 2.2 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 0.6 | GO:0006585 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.2 | 2.2 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 0.5 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.2 | 2.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.2 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 0.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.5 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.4 | GO:0099625 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 6.1 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 8.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 3.3 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 2.1 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 2.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 1.5 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 2.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 11.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 3.2 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.1 | 2.1 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.8 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.3 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.9 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 5.7 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.1 | 1.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 2.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 3.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.8 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cyclic nucleotide metabolic process(GO:0030801) positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) positive regulation of nucleotide biosynthetic process(GO:0030810) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of cyclase activity(GO:0031281) positive regulation of adenylate cyclase activity(GO:0045762) positive regulation of lyase activity(GO:0051349) positive regulation of purine nucleotide biosynthetic process(GO:1900373) |
| 0.1 | 1.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.5 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.7 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 3.4 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.1 | 3.0 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 2.6 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.8 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 3.0 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 1.8 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 11.8 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 6.7 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 0.3 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.6 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 13.4 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 1.6 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 1.3 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 1.3 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 3.8 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.8 | GO:0010469 | regulation of receptor activity(GO:0010469) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 15.1 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.8 | 20.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.5 | 6.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 7.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 5.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 6.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 2.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 5.2 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 11.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 0.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 3.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 3.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 2.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 2.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 7.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 7.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 2.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 13.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 2.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 7.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 2.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 10.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 7.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 6.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 3.0 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 3.7 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 1.2 | 6.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.2 | 15.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 1.0 | 11.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.8 | 2.4 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.7 | 6.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 8.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.6 | 1.8 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.6 | 4.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.6 | 3.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 19.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 11.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.4 | 1.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 2.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.3 | 2.4 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.3 | 1.9 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.3 | 6.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 1.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 1.4 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.3 | 5.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 3.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 0.7 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.2 | 3.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 4.5 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.2 | 4.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 0.6 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 2.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 2.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 2.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 4.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.5 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 8.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.6 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 11.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 3.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 16.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 16.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 2.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.9 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 1.0 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 7.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.9 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.8 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 11.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 1.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.5 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 15.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 5.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 2.5 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.3 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 1.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 10.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 8.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 12.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 2.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 3.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 1.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 1.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 5.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |