Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
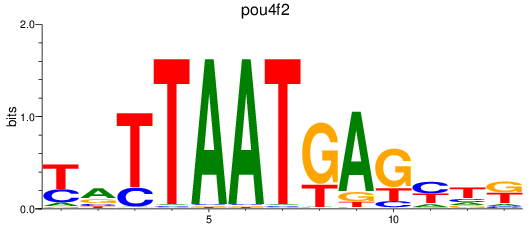
Results for pou4f2
Z-value: 1.47
Transcription factors associated with pou4f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou4f2
|
ENSDARG00000069737 | POU class 4 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou4f2 | dr11_v1_chr1_+_36436936_36436936 | 0.91 | 2.2e-37 | Click! |
Activity profile of pou4f2 motif
Sorted Z-values of pou4f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_15433671 | 35.56 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433518 | 34.77 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr2_-_21082695 | 30.84 |
ENSDART00000032502
|
nebl
|
nebulette |
| chr6_+_27146671 | 25.38 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr7_+_73630751 | 23.58 |
ENSDART00000159745
|
PCP4L1
|
si:dkey-46i9.1 |
| chr18_-_1185772 | 23.05 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr24_-_21923930 | 22.86 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr9_-_32753535 | 22.83 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr25_-_13842618 | 21.31 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr8_+_16025554 | 21.01 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr16_-_12173554 | 20.84 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr5_+_37966505 | 20.19 |
ENSDART00000127648
|
pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr4_-_1360495 | 19.48 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr19_-_32641725 | 19.25 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr19_-_5103313 | 17.56 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr7_+_25059845 | 17.02 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr12_-_35787801 | 16.27 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr1_-_50859053 | 15.75 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr2_-_30784198 | 15.71 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr19_-_31402429 | 15.47 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr11_-_37509001 | 14.60 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr14_-_31465905 | 13.86 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr16_-_26074529 | 13.45 |
ENSDART00000148653
ENSDART00000148923 |
tmem145
|
transmembrane protein 145 |
| chr16_+_29303971 | 13.07 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr17_+_8183393 | 12.90 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr20_-_18736281 | 12.84 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr19_-_5103141 | 12.65 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr6_+_54187643 | 11.92 |
ENSDART00000056830
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr18_-_40509006 | 11.71 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr13_+_36622100 | 11.59 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr18_+_783936 | 11.28 |
ENSDART00000193357
|
rpp25b
|
ribonuclease P and MRP subunit p25, b |
| chr2_+_50608099 | 10.83 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr20_+_41756996 | 10.69 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr21_+_28958471 | 10.53 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr7_+_25915089 | 10.52 |
ENSDART00000173648
|
hmgb3a
|
high mobility group box 3a |
| chr3_+_33341640 | 10.43 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr23_-_28141419 | 10.28 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr16_-_12173399 | 9.99 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr1_+_25801648 | 9.99 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr7_+_19552381 | 9.95 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr18_-_2433011 | 9.85 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr15_+_14826966 | 9.66 |
ENSDART00000157912
ENSDART00000171133 |
or130-1
|
odorant receptor, family H, subfamily 130, member 1 |
| chr13_-_14487524 | 9.45 |
ENSDART00000141103
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr1_-_53756851 | 9.06 |
ENSDART00000122445
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr13_+_1089942 | 9.00 |
ENSDART00000054322
|
cnrip1b
|
cannabinoid receptor interacting protein 1b |
| chr1_-_22512063 | 8.66 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr4_-_23908802 | 8.65 |
ENSDART00000138873
|
celf2
|
cugbp, Elav-like family member 2 |
| chr3_-_30434016 | 8.49 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr14_+_45883687 | 8.46 |
ENSDART00000114790
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr12_+_24952902 | 8.33 |
ENSDART00000189086
ENSDART00000014868 |
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr5_+_51443009 | 7.88 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr9_+_21793565 | 7.75 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr2_+_29249204 | 7.60 |
ENSDART00000168957
|
cdh18a
|
cadherin 18, type 2a |
| chr21_-_25612658 | 7.48 |
ENSDART00000115276
|
fibpb
|
fibroblast growth factor (acidic) intracellular binding protein b |
| chr4_-_1801519 | 7.40 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr15_-_16121496 | 7.31 |
ENSDART00000128624
|
sgk494a
|
uncharacterized serine/threonine-protein kinase SgK494a |
| chr17_+_11675362 | 7.05 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr16_+_25011994 | 7.03 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr2_+_12302527 | 6.91 |
ENSDART00000091298
ENSDART00000182569 |
arhgap21b
|
Rho GTPase activating protein 21b |
| chr18_+_34362608 | 6.73 |
ENSDART00000131478
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr23_-_15284757 | 6.60 |
ENSDART00000139135
|
sulf2b
|
sulfatase 2b |
| chr10_+_40284003 | 6.51 |
ENSDART00000062795
ENSDART00000193825 ENSDART00000113582 |
git2b
|
G protein-coupled receptor kinase interacting ArfGAP 2b |
| chr5_+_44064764 | 6.48 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr19_+_9459050 | 6.40 |
ENSDART00000186419
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr16_+_25107344 | 6.34 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
| chr14_+_25817628 | 6.33 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr12_+_32729470 | 5.55 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr2_-_40135942 | 5.52 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr11_+_14937904 | 5.43 |
ENSDART00000185103
|
CR550302.3
|
|
| chr9_-_43538328 | 5.35 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr13_+_31177934 | 5.21 |
ENSDART00000144568
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr8_+_44714336 | 5.04 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr22_+_10698549 | 4.88 |
ENSDART00000081228
|
abhd14a
|
abhydrolase domain containing 14A |
| chr6_-_40466861 | 4.77 |
ENSDART00000062724
|
crbn
|
cereblon |
| chr5_-_63515210 | 4.73 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr8_+_17884569 | 4.64 |
ENSDART00000134660
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr2_+_23006792 | 4.51 |
ENSDART00000027782
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr16_+_30301539 | 4.38 |
ENSDART00000186018
|
LO017848.1
|
|
| chr10_+_38775408 | 4.24 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr17_+_51499789 | 4.18 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr9_-_29003245 | 4.03 |
ENSDART00000183391
ENSDART00000188836 |
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr5_+_42381273 | 4.02 |
ENSDART00000142387
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr11_+_38280454 | 3.99 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr6_-_12172424 | 3.88 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr3_-_33574576 | 3.86 |
ENSDART00000184881
|
CR847537.1
|
|
| chr8_+_52314542 | 3.85 |
ENSDART00000013059
ENSDART00000125241 |
dbnlb
|
drebrin-like b |
| chr3_-_61362398 | 3.79 |
ENSDART00000156177
|
si:dkey-111k8.3
|
si:dkey-111k8.3 |
| chr7_-_33960170 | 3.67 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr2_+_16696052 | 3.47 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr15_+_45994123 | 3.46 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr13_+_31180084 | 3.34 |
ENSDART00000133774
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr14_+_28486213 | 3.23 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr9_+_29430432 | 3.15 |
ENSDART00000125632
|
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr24_+_19593197 | 3.10 |
ENSDART00000151923
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr11_-_42918971 | 2.75 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr13_-_16066997 | 2.74 |
ENSDART00000184790
|
SPATA48
|
spermatogenesis associated 48 |
| chr17_-_49438873 | 2.63 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr13_-_37109987 | 2.58 |
ENSDART00000136750
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr5_+_17624463 | 2.49 |
ENSDART00000183869
ENSDART00000081064 |
fbrsl1
|
fibrosin-like 1 |
| chr1_+_10318089 | 2.44 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr17_-_29902187 | 2.18 |
ENSDART00000009104
|
esrrb
|
estrogen-related receptor beta |
| chr7_+_53498152 | 2.16 |
ENSDART00000184497
|
znf609b
|
zinc finger protein 609b |
| chr2_-_3045861 | 2.14 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr9_+_38088331 | 2.08 |
ENSDART00000123749
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr3_-_32169754 | 2.01 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr3_-_30384353 | 1.81 |
ENSDART00000186832
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr15_-_5720583 | 1.73 |
ENSDART00000158034
ENSDART00000190332 ENSDART00000109599 |
urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr7_-_18416741 | 1.62 |
ENSDART00000097882
|
sstr1b
|
somatostatin receptor 1b |
| chr10_-_29831944 | 1.48 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr3_-_23406964 | 1.43 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr11_+_44502410 | 1.25 |
ENSDART00000172998
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr17_+_6765621 | 1.25 |
ENSDART00000156637
ENSDART00000007622 |
afg1la
|
AFG1 like ATPase a |
| chr21_+_36623162 | 1.24 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr7_+_32369026 | 1.17 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr3_+_56276346 | 1.17 |
ENSDART00000157770
|
PRKCA
|
si:ch73-374l24.1 |
| chr6_+_39184236 | 1.15 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr10_+_25947946 | 1.09 |
ENSDART00000064393
|
ufm1
|
ubiquitin-fold modifier 1 |
| chr1_+_47499888 | 1.06 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr25_+_6186823 | 0.96 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr7_-_17779644 | 0.89 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr4_+_11695979 | 0.81 |
ENSDART00000137736
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr14_-_7306983 | 0.73 |
ENSDART00000158914
|
si:ch211-51f19.1
|
si:ch211-51f19.1 |
| chr10_+_39084354 | 0.62 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr4_+_13428993 | 0.58 |
ENSDART00000067151
|
si:dkey-39a18.1
|
si:dkey-39a18.1 |
| chr20_+_13781779 | 0.54 |
ENSDART00000142999
ENSDART00000152471 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr2_-_9818640 | 0.53 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr21_+_20396858 | 0.48 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr13_-_42400647 | 0.44 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr14_-_32959851 | 0.40 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr5_-_24270989 | 0.34 |
ENSDART00000146251
|
si:ch211-137i24.12
|
si:ch211-137i24.12 |
| chr17_-_12926456 | 0.33 |
ENSDART00000044126
|
INSM2
|
INSM transcriptional repressor 2 |
| chr9_-_19699728 | 0.03 |
ENSDART00000166780
|
si:ch211-141e20.2
|
si:ch211-141e20.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou4f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 30.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 5.7 | 22.8 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 3.4 | 30.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 2.7 | 16.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 2.5 | 10.0 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 2.3 | 11.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 2.1 | 6.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 1.9 | 7.8 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 1.7 | 13.9 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 1.7 | 6.7 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 1.6 | 4.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 1.5 | 7.4 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 1.5 | 14.6 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 1.3 | 11.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 1.0 | 9.0 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 1.0 | 11.7 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.9 | 15.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.9 | 26.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.9 | 17.0 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.8 | 5.5 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.8 | 3.8 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.7 | 10.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.6 | 11.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.6 | 1.7 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.5 | 6.6 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.4 | 10.4 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.3 | 21.3 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.3 | 19.5 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.2 | 8.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 1.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 1.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 8.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 3.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 6.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.2 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.2 | 3.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 25.4 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.1 | 5.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 0.5 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 1.6 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.1 | 5.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 20.2 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.1 | 3.7 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 21.5 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.1 | 2.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 2.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 9.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.1 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 1.0 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 7.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.0 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 8.7 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 1.3 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 1.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 4.0 | GO:0006470 | protein dephosphorylation(GO:0006470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 10.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.6 | 11.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.5 | 14.6 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.7 | 31.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.7 | 4.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.6 | 13.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.6 | 6.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 17.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 32.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.4 | 1.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 11.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 17.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.2 | 30.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 39.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 6.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 6.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 17.8 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 6.6 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 0.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 3.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 5.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 14.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 13.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 31.1 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 19.3 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 30.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 4.7 | 70.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 2.1 | 10.4 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 1.8 | 10.5 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.6 | 4.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 1.5 | 7.4 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.9 | 17.0 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.8 | 20.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.8 | 14.6 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.8 | 26.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.8 | 25.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.7 | 6.3 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.7 | 8.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.6 | 3.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.6 | 10.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.4 | 2.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 6.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 13.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 6.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 7.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 7.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.3 | 5.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 6.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.3 | 2.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 1.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 19.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 3.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 4.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 7.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 2.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 6.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 3.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 7.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 18.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 17.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 9.0 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 39.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 4.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 6.8 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 17.9 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 37.7 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 8.5 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 4.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 19.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.8 | 20.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.7 | 10.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.4 | 15.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.4 | 11.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 13.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 20.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 1.4 | 17.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.9 | 13.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.8 | 10.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 16.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.3 | 6.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 3.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 7.8 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 2.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |