Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
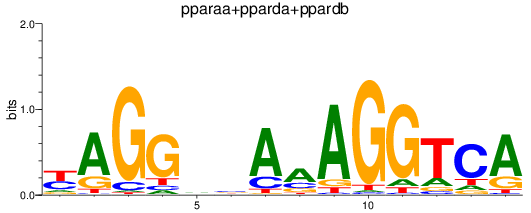
Results for pparaa+pparda+ppardb
Z-value: 0.85
Transcription factors associated with pparaa+pparda+ppardb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ppardb
|
ENSDARG00000009473 | peroxisome proliferator-activated receptor delta b |
|
pparaa
|
ENSDARG00000031777 | peroxisome proliferator-activated receptor alpha a |
|
pparda
|
ENSDARG00000044525 | peroxisome proliferator-activated receptor delta a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ppardb | dr11_v1_chr8_+_23711842_23711842 | -0.31 | 2.5e-03 | Click! |
| pparaa | dr11_v1_chr4_-_18635005_18635005 | -0.21 | 3.8e-02 | Click! |
| pparda | dr11_v1_chr22_+_1006573_1006573 | -0.10 | 3.2e-01 | Click! |
Activity profile of pparaa+pparda+ppardb motif
Sorted Z-values of pparaa+pparda+ppardb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_11201096 | 11.64 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr7_+_34297271 | 9.30 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr6_-_39764995 | 9.28 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr6_-_39765546 | 8.86 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr21_-_19006631 | 8.02 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr9_-_43082945 | 7.58 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr7_+_34296789 | 7.07 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr19_+_7043634 | 6.59 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr3_+_27770110 | 6.06 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr20_+_15015557 | 5.34 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr10_+_35417099 | 5.21 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr20_+_47953047 | 5.02 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr17_-_32370047 | 4.79 |
ENSDART00000145487
|
klf11b
|
Kruppel-like factor 11b |
| chr23_+_17417539 | 4.61 |
ENSDART00000182605
|
BX649300.2
|
|
| chr14_-_48103207 | 4.58 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr21_+_15704556 | 4.37 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr20_-_47953524 | 4.26 |
ENSDART00000167986
|
HADHB
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr15_+_404891 | 4.13 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr18_-_48508585 | 3.82 |
ENSDART00000133364
|
kcnj1a.4
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 4 |
| chr1_+_40802454 | 3.72 |
ENSDART00000193568
|
cpz
|
carboxypeptidase Z |
| chr5_-_30984010 | 3.36 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr6_+_60036767 | 3.34 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr19_+_40861853 | 3.33 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr19_+_2631565 | 3.29 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr8_+_999421 | 3.27 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr16_-_50229193 | 3.19 |
ENSDART00000161782
ENSDART00000010081 |
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr7_-_24046999 | 3.15 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr3_-_34561624 | 3.10 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr14_-_29826659 | 3.07 |
ENSDART00000138413
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr3_+_30257582 | 2.92 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr20_-_34164278 | 2.78 |
ENSDART00000153128
|
hmcn1
|
hemicentin 1 |
| chr18_-_48517040 | 2.77 |
ENSDART00000143645
|
kcnj1a.3
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 3 |
| chr17_+_30546579 | 2.70 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr3_+_58167288 | 2.66 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr8_-_2529878 | 2.64 |
ENSDART00000056767
|
acaa2
|
acetyl-CoA acyltransferase 2 |
| chr22_-_7050 | 2.47 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr3_-_18805225 | 2.46 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr20_+_43379029 | 2.33 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr22_+_22888 | 2.30 |
ENSDART00000082471
|
mfap2
|
microfibril associated protein 2 |
| chr22_+_18469004 | 2.29 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr17_+_30545895 | 2.28 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr24_+_2961098 | 2.26 |
ENSDART00000163760
ENSDART00000170835 |
eci2
|
enoyl-CoA delta isomerase 2 |
| chr13_+_9842276 | 2.26 |
ENSDART00000041700
ENSDART00000141406 |
prdx3
|
peroxiredoxin 3 |
| chr15_+_28268135 | 2.21 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr24_+_81527 | 2.20 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr18_+_13315739 | 2.15 |
ENSDART00000143404
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr16_-_30563129 | 2.11 |
ENSDART00000191716
|
lmna
|
lamin A |
| chr23_-_39636195 | 2.07 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr3_-_53486169 | 2.07 |
ENSDART00000115243
|
soul5
|
heme-binding protein soul5 |
| chr1_-_17797802 | 2.07 |
ENSDART00000041215
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr10_+_11265387 | 1.95 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr19_-_7043355 | 1.90 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr1_+_58139102 | 1.90 |
ENSDART00000134826
|
si:ch211-15j1.4
|
si:ch211-15j1.4 |
| chr8_-_979735 | 1.88 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr1_-_52292235 | 1.82 |
ENSDART00000132638
|
si:dkey-121b10.7
|
si:dkey-121b10.7 |
| chr9_+_41224100 | 1.81 |
ENSDART00000141792
|
stat1b
|
signal transducer and activator of transcription 1b |
| chr2_-_26476030 | 1.77 |
ENSDART00000145262
ENSDART00000132125 |
acadm
|
acyl-CoA dehydrogenase medium chain |
| chr9_+_30626416 | 1.74 |
ENSDART00000147813
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr18_+_16744307 | 1.73 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr20_-_48604199 | 1.62 |
ENSDART00000161762
ENSDART00000170894 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr23_-_16737161 | 1.61 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr20_-_26937453 | 1.60 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr15_+_31820536 | 1.58 |
ENSDART00000045921
|
frya
|
furry homolog a (Drosophila) |
| chr8_+_2530065 | 1.57 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr5_-_30984271 | 1.54 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr12_-_684200 | 1.54 |
ENSDART00000152122
|
si:ch211-176g6.2
|
si:ch211-176g6.2 |
| chr5_+_43870389 | 1.50 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr7_+_48675347 | 1.47 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr20_-_48604621 | 1.46 |
ENSDART00000161769
ENSDART00000157871 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr11_-_6974022 | 1.38 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr15_-_6946286 | 1.35 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr5_-_66301142 | 1.33 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr1_-_58868306 | 1.33 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr25_+_5288665 | 1.32 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr24_+_9590188 | 1.29 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr23_-_16734009 | 1.24 |
ENSDART00000125449
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr17_-_12196865 | 1.24 |
ENSDART00000154694
|
kif28
|
kinesin family member 28 |
| chr20_-_26936887 | 1.23 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr3_-_58116314 | 1.22 |
ENSDART00000154901
|
si:ch211-256e16.6
|
si:ch211-256e16.6 |
| chr1_+_57050899 | 1.16 |
ENSDART00000152601
|
si:ch211-1f22.14
|
si:ch211-1f22.14 |
| chr1_+_58353661 | 1.12 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr1_-_35113974 | 1.12 |
ENSDART00000192811
ENSDART00000167461 |
BX897691.1
|
|
| chr4_+_60492313 | 1.12 |
ENSDART00000191043
ENSDART00000191631 |
si:dkey-211i20.2
|
si:dkey-211i20.2 |
| chr1_-_46859398 | 1.11 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr7_+_17356482 | 1.09 |
ENSDART00000063629
|
nitr3r.1l
|
novel immune-type receptor 3, related 1-like |
| chr1_+_58260886 | 1.05 |
ENSDART00000110453
|
si:dkey-222h21.8
|
si:dkey-222h21.8 |
| chr8_-_20291922 | 1.02 |
ENSDART00000148304
|
myo1f
|
myosin IF |
| chr21_+_26720803 | 0.99 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr5_-_323712 | 0.98 |
ENSDART00000188793
|
HOOK3
|
hook microtubule tethering protein 3 |
| chr18_+_27926839 | 0.91 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr3_+_1179601 | 0.82 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr7_+_11459235 | 0.78 |
ENSDART00000159611
|
il16
|
interleukin 16 |
| chr11_+_45153104 | 0.75 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr2_+_7213146 | 0.75 |
ENSDART00000191134
ENSDART00000146547 |
si:ch211-13f8.2
|
si:ch211-13f8.2 |
| chr5_-_41875834 | 0.71 |
ENSDART00000133592
|
si:dkey-65b12.10
|
si:dkey-65b12.10 |
| chr25_+_36347126 | 0.71 |
ENSDART00000152449
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr22_+_9287929 | 0.70 |
ENSDART00000193522
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr9_-_1939232 | 0.70 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr4_+_75577480 | 0.68 |
ENSDART00000188196
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr22_+_9114416 | 0.67 |
ENSDART00000190169
|
nlrp15
|
NACHT, LRR and PYD domains-containing protein 15 |
| chr17_+_6538733 | 0.66 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr3_+_32546499 | 0.66 |
ENSDART00000151739
|
pax10
|
paired box 10 |
| chr4_+_33547662 | 0.66 |
ENSDART00000150439
|
si:dkey-84h14.2
|
si:dkey-84h14.2 |
| chr13_+_43050562 | 0.65 |
ENSDART00000016602
|
cdh23
|
cadherin-related 23 |
| chr17_-_20711735 | 0.62 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr4_-_991043 | 0.60 |
ENSDART00000184706
|
naga
|
N-acetylgalactosaminidase, alpha |
| chr4_-_36032177 | 0.59 |
ENSDART00000170143
|
zgc:174180
|
zgc:174180 |
| chr22_+_9239831 | 0.58 |
ENSDART00000133720
|
si:ch211-250k18.5
|
si:ch211-250k18.5 |
| chr4_-_50926767 | 0.56 |
ENSDART00000183430
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr22_+_10163901 | 0.55 |
ENSDART00000190468
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr22_-_10774735 | 0.55 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr14_+_24258562 | 0.52 |
ENSDART00000106074
ENSDART00000173227 |
chrm1a
|
cholinergic receptor, muscarinic 1a |
| chr23_-_24488696 | 0.51 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr25_-_35113891 | 0.50 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr8_-_8698607 | 0.50 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr13_+_829585 | 0.48 |
ENSDART00000029051
|
gsta.2
|
glutathione S-transferase, alpha tandem duplicate 2 |
| chr7_+_23940933 | 0.46 |
ENSDART00000173628
|
si:dkey-183c6.7
|
si:dkey-183c6.7 |
| chr16_-_25680666 | 0.41 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr1_+_58442694 | 0.39 |
ENSDART00000160897
|
zgc:194906
|
zgc:194906 |
| chr7_+_17353030 | 0.38 |
ENSDART00000098020
|
si:busm1-169i8.11
|
si:busm1-169i8.11 |
| chr2_-_56649883 | 0.37 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr15_+_45640906 | 0.36 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr13_-_28688104 | 0.36 |
ENSDART00000133827
|
pcgf6
|
polycomb group ring finger 6 |
| chr18_-_3456117 | 0.34 |
ENSDART00000158508
|
myo7aa
|
myosin VIIAa |
| chr10_-_44027391 | 0.33 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr6_-_35046735 | 0.32 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr22_-_38274188 | 0.31 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr6_-_28117995 | 0.30 |
ENSDART00000147253
|
si:ch73-194h10.3
|
si:ch73-194h10.3 |
| chr15_-_12500938 | 0.28 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr20_+_46286082 | 0.27 |
ENSDART00000073519
|
taar14f
|
trace amine associated receptor 14f |
| chr12_-_5120175 | 0.24 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr11_-_44999858 | 0.21 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr19_+_32158010 | 0.20 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr9_+_32069989 | 0.19 |
ENSDART00000139540
|
si:dkey-83m22.7
|
si:dkey-83m22.7 |
| chr12_-_44199316 | 0.17 |
ENSDART00000170378
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr22_+_9069081 | 0.16 |
ENSDART00000187842
|
BX248395.1
|
|
| chr5_+_38200807 | 0.15 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr5_-_36549024 | 0.14 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr23_+_44634187 | 0.13 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr15_+_17030941 | 0.12 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr12_+_26670778 | 0.11 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr4_-_16451375 | 0.10 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr2_-_42960353 | 0.09 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr20_-_38836161 | 0.07 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr5_-_41860556 | 0.03 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pparaa+pparda+ppardb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 16.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.6 | 18.1 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.7 | 2.2 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.5 | 5.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.4 | 15.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.4 | 2.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.3 | 1.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 1.0 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.3 | 5.3 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.3 | 8.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 0.7 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 1.9 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 0.7 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 7.6 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.2 | 3.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 0.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 1.8 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 1.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.7 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 4.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.8 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.6 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.1 | 2.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 6.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 1.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 2.8 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 3.6 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 2.2 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.1 | 4.6 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 11.3 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 3.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.8 | GO:0050930 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 3.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.2 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 2.3 | GO:0048048 | embryonic eye morphogenesis(GO:0048048) |
| 0.0 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 3.7 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 4.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 6.4 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 18.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.6 | 11.6 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 2.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 5.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 4.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 3.1 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 11.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 3.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 13.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 27.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 2.7 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 1.7 | GO:0044420 | extracellular matrix component(GO:0044420) |
| 0.0 | 3.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.7 | 5.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.6 | 18.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 1.6 | 8.0 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 1.5 | 4.6 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 1.2 | 6.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.9 | 2.6 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.6 | 2.5 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.5 | 6.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 5.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.3 | 1.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 2.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 1.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.6 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.2 | 0.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 2.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 3.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.7 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 3.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 0.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 1.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 3.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 2.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 3.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 2.2 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 3.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 1.5 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 1.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.5 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 11.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 3.1 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.1 | GO:0020037 | heme binding(GO:0020037) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 10.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 12.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 8.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 7.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 16.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |