Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
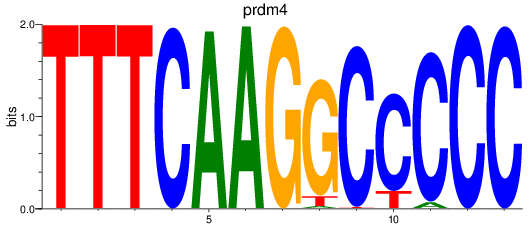
Results for prdm4
Z-value: 0.37
Transcription factors associated with prdm4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm4
|
ENSDARG00000017366 | PR domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prdm4 | dr11_v1_chr4_-_14926637_14926637 | 0.01 | 9.5e-01 | Click! |
Activity profile of prdm4 motif
Sorted Z-values of prdm4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_31621774 | 5.92 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr9_-_13355071 | 3.60 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr23_-_24682244 | 2.68 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr9_+_41690153 | 1.94 |
ENSDART00000100226
|
ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr6_-_9707599 | 1.49 |
ENSDART00000039280
|
fzd7b
|
frizzled class receptor 7b |
| chr5_+_27442247 | 1.48 |
ENSDART00000184129
|
loxl2b
|
lysyl oxidase-like 2b |
| chr10_-_9115383 | 1.09 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr22_-_21046843 | 0.97 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr19_+_2670130 | 0.94 |
ENSDART00000152152
|
si:ch73-24k9.2
|
si:ch73-24k9.2 |
| chr9_+_22359919 | 0.80 |
ENSDART00000009591
|
crygs4
|
crystallin, gamma S4 |
| chr10_+_43037064 | 0.78 |
ENSDART00000160159
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr21_-_37027252 | 0.78 |
ENSDART00000076483
|
zgc:77151
|
zgc:77151 |
| chr4_-_2868112 | 0.69 |
ENSDART00000133843
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr3_+_25999477 | 0.65 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chrM_+_9735 | 0.65 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr11_-_6868474 | 0.62 |
ENSDART00000168372
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_+_12348213 | 0.55 |
ENSDART00000144920
ENSDART00000138759 ENSDART00000067082 |
clta
|
clathrin, light chain A |
| chr11_-_6868287 | 0.54 |
ENSDART00000037824
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr4_-_2867461 | 0.46 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr17_-_31611692 | 0.42 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr1_+_56180416 | 0.37 |
ENSDART00000089358
|
crb3b
|
crumbs homolog 3b |
| chr17_+_31611854 | 0.37 |
ENSDART00000011706
|
adprhl2
|
ADP-ribosylhydrolase like 2 |
| chr22_-_21046654 | 0.25 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr24_-_27409599 | 0.22 |
ENSDART00000041770
|
ccl34b.3
|
chemokine (C-C motif) ligand 34b, duplicate 3 |
| chr10_-_3332362 | 0.17 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr24_+_29912509 | 0.17 |
ENSDART00000168422
|
frrs1b
|
ferric-chelate reductase 1b |
| chr3_+_40407352 | 0.03 |
ENSDART00000155112
|
tnrc18
|
trinucleotide repeat containing 18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.7 | 3.6 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.4 | 1.5 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.2 | 1.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 0.7 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.5 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) clathrin coat of coated pit(GO:0030132) |
| 0.0 | 5.1 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.5 | 4.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 0.8 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 1.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.7 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 5.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |