Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
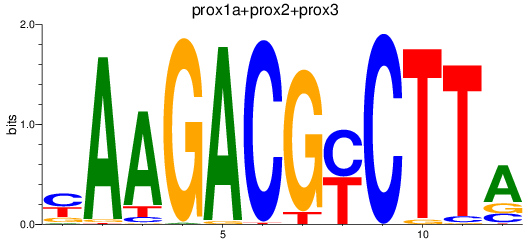
Results for prox1a+prox2+prox3
Z-value: 0.81
Transcription factors associated with prox1a+prox2+prox3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prox2
|
ENSDARG00000041952 | prospero homeobox 2 |
|
prox1a
|
ENSDARG00000055158 | prospero homeobox 1a |
|
prox3
|
ENSDARG00000088810 | prospero homeobox 3 |
|
prox3
|
ENSDARG00000113014 | prospero homeobox 3 |
|
prox2
|
ENSDARG00000117137 | prospero homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prox1a | dr11_v1_chr17_-_32865788_32865788 | -0.73 | 6.3e-17 | Click! |
| prox1b | dr11_v1_chr7_-_19940473_19940473 | -0.23 | 2.5e-02 | Click! |
| prox2 | dr11_v1_chr17_-_52521002_52521002 | 0.00 | 9.9e-01 | Click! |
Activity profile of prox1a+prox2+prox3 motif
Sorted Z-values of prox1a+prox2+prox3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_29952169 | 25.00 |
ENSDART00000173540
ENSDART00000173940 ENSDART00000173906 ENSDART00000173772 ENSDART00000173506 ENSDART00000039657 |
tpma
|
alpha-tropomyosin |
| chr7_+_29951997 | 22.03 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr2_+_2470687 | 14.45 |
ENSDART00000184024
ENSDART00000061955 |
myl13
|
myosin, light chain 13 |
| chr17_-_5583345 | 12.46 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr19_-_31042570 | 7.84 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr15_+_32711663 | 7.74 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr25_+_19947096 | 6.53 |
ENSDART00000154221
|
kcna6a
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 a |
| chr9_-_42696408 | 6.28 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr25_+_19947298 | 6.15 |
ENSDART00000067648
|
kcna6a
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 a |
| chr19_+_42886413 | 5.41 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr23_-_5759242 | 4.83 |
ENSDART00000055087
|
phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr5_+_37517800 | 4.59 |
ENSDART00000048107
|
FP102018.1
|
Danio rerio latent transforming growth factor beta binding protein 3 (ltbp3), mRNA. |
| chr5_+_37785152 | 4.36 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr11_-_24681292 | 4.13 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr5_+_27440325 | 3.97 |
ENSDART00000185815
ENSDART00000144013 |
loxl2b
|
lysyl oxidase-like 2b |
| chr9_-_23033818 | 3.80 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr5_+_13870340 | 3.48 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr18_+_7204378 | 3.32 |
ENSDART00000142905
|
vwf
|
von Willebrand factor |
| chr14_+_31751260 | 3.08 |
ENSDART00000169796
|
si:dkeyp-11e3.1
|
si:dkeyp-11e3.1 |
| chr21_-_303766 | 2.61 |
ENSDART00000150913
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr8_-_47800754 | 2.50 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr3_-_59981162 | 2.14 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr23_+_16889352 | 2.07 |
ENSDART00000145275
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr10_-_8294965 | 2.06 |
ENSDART00000167380
|
plpp1a
|
phospholipid phosphatase 1a |
| chr13_+_7442023 | 2.02 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr4_+_77973876 | 2.01 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr10_-_8295294 | 2.00 |
ENSDART00000075412
ENSDART00000163803 |
plpp1a
|
phospholipid phosphatase 1a |
| chr8_-_4475908 | 1.96 |
ENSDART00000191027
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr9_+_8968702 | 1.95 |
ENSDART00000008490
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr1_-_59065217 | 1.95 |
ENSDART00000188972
|
CABZ01080042.1
|
|
| chr15_+_37954666 | 1.90 |
ENSDART00000126534
ENSDART00000155548 |
si:dkey-238d18.7
|
si:dkey-238d18.7 |
| chr8_+_25616946 | 1.83 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr23_+_21663631 | 1.78 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr21_-_22831388 | 1.77 |
ENSDART00000151040
|
angptl5
|
angiopoietin-like 5 |
| chr15_-_29162193 | 1.75 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr16_+_42772678 | 1.73 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr1_+_6161930 | 1.65 |
ENSDART00000037036
|
cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr15_-_23475051 | 1.62 |
ENSDART00000152460
|
nlrx1
|
NLR family member X1 |
| chr17_-_21832119 | 1.60 |
ENSDART00000153539
ENSDART00000079008 |
si:ch211-208g24.8
|
si:ch211-208g24.8 |
| chr5_-_61638125 | 1.60 |
ENSDART00000134314
|
si:dkey-261j4.3
|
si:dkey-261j4.3 |
| chr2_+_6885852 | 1.59 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr12_-_23365737 | 1.55 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr20_-_54014373 | 1.47 |
ENSDART00000152934
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr16_-_54919260 | 1.44 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr12_+_27117609 | 1.40 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr20_+_192170 | 1.38 |
ENSDART00000189675
|
cx28.8
|
connexin 28.8 |
| chr8_-_49345388 | 1.38 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr25_+_8407892 | 1.31 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr7_-_25934540 | 1.21 |
ENSDART00000192761
|
BX005022.2
|
|
| chr5_+_42136359 | 1.20 |
ENSDART00000083731
|
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr17_+_48164536 | 1.17 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr3_+_11072958 | 1.16 |
ENSDART00000158899
|
mrps7
|
mitochondrial ribosomal protein S7 |
| chr12_-_4408828 | 1.08 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr19_-_22621811 | 1.06 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr14_+_24840669 | 1.00 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr13_-_27384697 | 0.98 |
ENSDART00000146230
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr12_-_18898413 | 0.96 |
ENSDART00000181281
ENSDART00000121866 |
desi1b
|
desumoylating isopeptidase 1b |
| chr4_+_30788743 | 0.94 |
ENSDART00000184557
|
si:dkey-178j11.5
|
si:dkey-178j11.5 |
| chr7_+_19562045 | 0.86 |
ENSDART00000077545
|
slc7a7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr9_+_38684871 | 0.86 |
ENSDART00000147938
ENSDART00000004462 |
heg1
|
heart development protein with EGF-like domains 1 |
| chr14_-_28566238 | 0.85 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr20_-_14924858 | 0.84 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr19_-_22621991 | 0.78 |
ENSDART00000175106
|
pleca
|
plectin a |
| chr16_+_11779761 | 0.77 |
ENSDART00000140297
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr15_+_2190229 | 0.75 |
ENSDART00000147710
|
rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr12_+_6065661 | 0.75 |
ENSDART00000142418
|
sgms1
|
sphingomyelin synthase 1 |
| chr13_+_47050726 | 0.73 |
ENSDART00000140045
|
anapc1
|
anaphase promoting complex subunit 1 |
| chr5_-_51484156 | 0.71 |
ENSDART00000162064
|
CR388055.1
|
|
| chr5_+_22510639 | 0.70 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr17_-_106291 | 0.70 |
ENSDART00000160738
|
arhgap11a
|
Rho GTPase activating protein 11A |
| chr6_-_34006917 | 0.65 |
ENSDART00000190702
ENSDART00000184003 |
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr22_+_9287929 | 0.65 |
ENSDART00000193522
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr15_-_31177324 | 0.64 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr19_+_7929704 | 0.58 |
ENSDART00000147015
|
si:dkey-266f7.4
|
si:dkey-266f7.4 |
| chr3_+_22335030 | 0.56 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr25_-_10630496 | 0.56 |
ENSDART00000153639
ENSDART00000181722 ENSDART00000177834 |
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr4_-_50933168 | 0.53 |
ENSDART00000150296
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr4_-_75899294 | 0.45 |
ENSDART00000157887
|
si:dkey-261j11.3
|
si:dkey-261j11.3 |
| chr14_-_30971264 | 0.44 |
ENSDART00000010512
|
zgc:92907
|
zgc:92907 |
| chrM_+_9052 | 0.44 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr4_+_47789703 | 0.43 |
ENSDART00000161323
|
si:ch211-196h24.2
|
si:ch211-196h24.2 |
| chr9_+_1138802 | 0.42 |
ENSDART00000191130
ENSDART00000187308 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr23_+_35672542 | 0.41 |
ENSDART00000046268
|
pmelb
|
premelanosome protein b |
| chr18_+_33264609 | 0.41 |
ENSDART00000050639
|
v2ra20
|
vomeronasal 2 receptor, a20 |
| chr9_+_1138323 | 0.39 |
ENSDART00000190352
ENSDART00000190387 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr10_+_6697369 | 0.38 |
ENSDART00000148606
|
si:ch211-57m13.8
|
si:ch211-57m13.8 |
| chr22_+_9239831 | 0.27 |
ENSDART00000133720
|
si:ch211-250k18.5
|
si:ch211-250k18.5 |
| chr7_+_6941583 | 0.24 |
ENSDART00000160709
ENSDART00000157634 |
rbm14b
|
RNA binding motif protein 14b |
| chr14_+_9287683 | 0.23 |
ENSDART00000122485
|
msnb
|
moesin b |
| chr4_+_5196469 | 0.17 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr9_-_53817143 | 0.17 |
ENSDART00000148764
ENSDART00000075537 |
ednrbb
|
endothelin receptor type Bb |
| chr1_-_9485939 | 0.17 |
ENSDART00000157814
|
micall2b
|
mical-like 2b |
| chr14_+_28473173 | 0.13 |
ENSDART00000017075
|
xiap
|
X-linked inhibitor of apoptosis |
| chr6_+_11250033 | 0.10 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr1_-_9486214 | 0.09 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr10_+_43037064 | 0.06 |
ENSDART00000160159
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr4_-_71551644 | 0.02 |
ENSDART00000168064
|
si:dkey-27n6.1
|
si:dkey-27n6.1 |
| chr4_+_3455665 | 0.01 |
ENSDART00000058277
|
znf800b
|
zinc finger protein 800b |
| chr13_-_31866276 | 0.00 |
ENSDART00000140164
|
syt14a
|
synaptotagmin XIVa |
Network of associatons between targets according to the STRING database.
First level regulatory network of prox1a+prox2+prox3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.5 | 2.0 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.5 | 1.8 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.5 | 47.9 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.4 | 4.8 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.4 | 7.7 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.3 | 2.6 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.3 | 1.4 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 3.5 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.2 | 2.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 2.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 12.5 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 0.8 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 2.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 10.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 4.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 1.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 4.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 2.0 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 3.3 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 3.8 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 1.7 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 2.5 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 6.3 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.7 | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle(GO:0007091) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.1 | 4.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.4 | 2.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 46.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 12.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 13.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 2.0 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 4.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 2.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 16.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 1.4 | 4.1 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.6 | 3.5 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 45.8 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.3 | 13.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 4.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 1.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 12.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 0.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.8 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 1.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 4.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 6.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 7.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 1.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 2.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.5 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 12.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 3.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |