Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
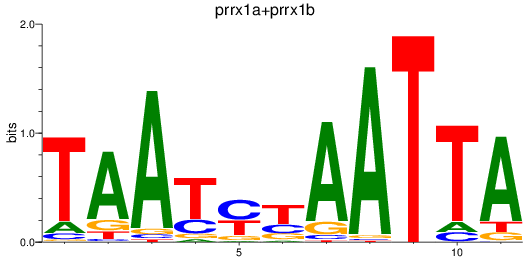
Results for prrx1a+prrx1b
Z-value: 1.10
Transcription factors associated with prrx1a+prrx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prrx1a
|
ENSDARG00000033971 | paired related homeobox 1a |
|
prrx1b
|
ENSDARG00000042027 | paired related homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prrx1b | dr11_v1_chr20_+_34512130_34512130 | -0.54 | 2.4e-08 | Click! |
| prrx1a | dr11_v1_chr2_-_23172708_23172708 | -0.41 | 3.3e-05 | Click! |
Activity profile of prrx1a+prrx1b motif
Sorted Z-values of prrx1a+prrx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 37.84 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr19_-_3167729 | 10.89 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr25_+_31267268 | 9.80 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr25_-_13381854 | 7.16 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr23_-_24343363 | 7.10 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr25_+_31277415 | 7.06 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr2_+_38039857 | 6.96 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr15_-_16098531 | 6.77 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr4_+_9669717 | 6.71 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr21_+_27382893 | 6.69 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr24_-_38079261 | 6.68 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr12_+_2428247 | 6.66 |
ENSDART00000152529
|
lrrc18b
|
leucine rich repeat containing 18b |
| chr6_+_23887314 | 6.60 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr13_+_24279021 | 6.30 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr18_-_47662696 | 6.22 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr1_+_17676745 | 6.05 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr6_+_40354424 | 5.92 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr12_-_558201 | 5.82 |
ENSDART00000168586
ENSDART00000158355 |
bsk146
|
brain specific kinase 146 |
| chr11_-_37509001 | 5.57 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr1_+_25801648 | 5.25 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr8_+_3820134 | 5.18 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr4_-_14315855 | 5.07 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr11_+_36243774 | 5.03 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr9_-_48397702 | 4.84 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr5_-_31901468 | 4.81 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr25_+_26921480 | 4.63 |
ENSDART00000155949
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr2_+_30916188 | 4.52 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr14_+_19258702 | 4.44 |
ENSDART00000187087
ENSDART00000005738 |
slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr13_-_29421331 | 4.43 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr18_-_21271373 | 4.41 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr19_+_31771270 | 4.39 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr15_-_44512461 | 4.38 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr21_+_13861589 | 4.36 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr12_+_18681477 | 4.32 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr17_-_14726824 | 4.00 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr21_+_3093419 | 3.98 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr20_-_27225876 | 3.97 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr8_-_53195500 | 3.96 |
ENSDART00000132000
|
gabrd
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr21_+_19008168 | 3.92 |
ENSDART00000136196
ENSDART00000128381 ENSDART00000176624 |
nefla
|
neurofilament, light polypeptide a |
| chr18_-_26785861 | 3.90 |
ENSDART00000098361
|
nmba
|
neuromedin Ba |
| chr20_-_9462433 | 3.86 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr22_+_20208185 | 3.72 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr2_-_39558643 | 3.69 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr17_-_15657029 | 3.68 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr10_-_26744131 | 3.66 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr16_-_16590780 | 3.66 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr2_+_23823622 | 3.64 |
ENSDART00000099581
|
si:dkey-24c2.9
|
si:dkey-24c2.9 |
| chr18_+_1703984 | 3.61 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr21_-_27881752 | 3.60 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr16_-_27640995 | 3.60 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr22_-_32507966 | 3.58 |
ENSDART00000104693
|
pcbp4
|
poly(rC) binding protein 4 |
| chr25_+_22730490 | 3.58 |
ENSDART00000149455
|
abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr1_-_45553602 | 3.57 |
ENSDART00000143664
|
grin2bb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b |
| chr1_+_18811679 | 3.56 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr25_+_29160102 | 3.56 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr23_-_12345764 | 3.56 |
ENSDART00000133956
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr1_-_18811517 | 3.49 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr21_+_13366353 | 3.45 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr23_+_45584223 | 3.40 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr17_-_37214196 | 3.40 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr8_+_41647539 | 3.28 |
ENSDART00000136492
ENSDART00000138799 ENSDART00000134404 |
si:ch211-158d24.4
|
si:ch211-158d24.4 |
| chr1_-_10647484 | 3.28 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr6_+_49412754 | 3.25 |
ENSDART00000027398
|
kcna2a
|
potassium voltage-gated channel, shaker-related subfamily, member 2a |
| chr10_+_3299829 | 3.24 |
ENSDART00000183684
|
zgc:56235
|
zgc:56235 |
| chr24_-_38657683 | 3.20 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr9_-_48407408 | 3.17 |
ENSDART00000058248
|
zgc:172182
|
zgc:172182 |
| chr15_+_22435460 | 3.09 |
ENSDART00000031976
|
tmem136a
|
transmembrane protein 136a |
| chr23_+_26026383 | 3.08 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr24_-_7697274 | 3.07 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr10_-_34772211 | 3.06 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr19_-_25113660 | 3.06 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr7_-_69983462 | 3.03 |
ENSDART00000123380
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr17_-_16965809 | 3.01 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr12_-_47601845 | 3.00 |
ENSDART00000169548
ENSDART00000182889 |
rgs7b
|
regulator of G protein signaling 7b |
| chr9_+_29603649 | 2.95 |
ENSDART00000140477
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr16_-_42872571 | 2.92 |
ENSDART00000154757
ENSDART00000102345 |
txnipb
|
thioredoxin interacting protein b |
| chr24_-_38384432 | 2.91 |
ENSDART00000140739
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr8_-_7391721 | 2.90 |
ENSDART00000149836
|
lhfpl4b
|
LHFPL tetraspan subfamily member 4b |
| chr3_-_55404985 | 2.89 |
ENSDART00000154274
|
mafga
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ga |
| chr9_+_13733468 | 2.88 |
ENSDART00000165954
ENSDART00000160355 ENSDART00000081040 ENSDART00000138254 ENSDART00000081015 ENSDART00000141314 |
abi2a
|
abl-interactor 2a |
| chr5_+_37966505 | 2.81 |
ENSDART00000127648
|
pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr22_+_18389271 | 2.81 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr14_-_49063157 | 2.77 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr7_-_52334840 | 2.76 |
ENSDART00000174173
|
CR938716.1
|
|
| chr17_-_26926577 | 2.74 |
ENSDART00000050202
|
rcan3
|
regulator of calcineurin 3 |
| chr25_+_33002963 | 2.69 |
ENSDART00000187366
|
CABZ01046980.1
|
|
| chr2_+_38554260 | 2.68 |
ENSDART00000171527
|
cdh24b
|
cadherin 24, type 2b |
| chr20_-_45423498 | 2.66 |
ENSDART00000098424
|
trib2
|
tribbles pseudokinase 2 |
| chr2_-_9489611 | 2.66 |
ENSDART00000146715
|
st6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr9_-_394088 | 2.64 |
ENSDART00000169014
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr12_-_32013125 | 2.64 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr1_+_10018466 | 2.60 |
ENSDART00000113551
|
trim2b
|
tripartite motif containing 2b |
| chr19_-_28130658 | 2.59 |
ENSDART00000079114
|
irx1b
|
iroquois homeobox 1b |
| chr6_+_55285578 | 2.57 |
ENSDART00000180183
|
zgc:109913
|
zgc:109913 |
| chr21_-_43022048 | 2.57 |
ENSDART00000138329
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr16_+_1100559 | 2.55 |
ENSDART00000092657
|
adamts16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr4_-_23963838 | 2.53 |
ENSDART00000133433
ENSDART00000132615 ENSDART00000135942 ENSDART00000139439 |
celf2
|
cugbp, Elav-like family member 2 |
| chr5_+_51079504 | 2.53 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr20_+_34717403 | 2.51 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr24_+_22485710 | 2.50 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr19_-_5103313 | 2.47 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr23_-_18913032 | 2.45 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr4_-_5019113 | 2.43 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr20_+_31287356 | 2.40 |
ENSDART00000007688
|
slc22a16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr6_+_6924637 | 2.40 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr6_+_32882821 | 2.40 |
ENSDART00000075780
|
ghrhrb
|
growth hormone releasing hormone receptor b |
| chr1_-_49250490 | 2.35 |
ENSDART00000150386
|
si:ch73-6k14.2
|
si:ch73-6k14.2 |
| chr19_-_41371978 | 2.29 |
ENSDART00000166063
ENSDART00000170343 |
slc25a13
|
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr6_+_58698475 | 2.29 |
ENSDART00000056138
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr22_-_15562933 | 2.29 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr23_-_44466257 | 2.27 |
ENSDART00000150126
|
si:ch1073-228j22.2
|
si:ch1073-228j22.2 |
| chr13_+_18331509 | 2.27 |
ENSDART00000181918
|
zgc:110319
|
zgc:110319 |
| chr24_-_17067284 | 2.27 |
ENSDART00000111237
|
armc3
|
armadillo repeat containing 3 |
| chr10_-_10607118 | 2.26 |
ENSDART00000101089
|
dbh
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr16_+_22654481 | 2.23 |
ENSDART00000179762
|
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr24_-_21923930 | 2.18 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr23_-_18707418 | 2.17 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr7_-_17028015 | 2.17 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr19_-_3240605 | 2.16 |
ENSDART00000105168
|
si:ch211-133n4.4
|
si:ch211-133n4.4 |
| chr5_+_40299568 | 2.15 |
ENSDART00000142157
|
arl15a
|
ADP-ribosylation factor-like 15a |
| chr10_-_34741738 | 2.14 |
ENSDART00000163072
|
dclk1a
|
doublecortin-like kinase 1a |
| chr2_-_50966124 | 2.14 |
ENSDART00000170470
|
ghrhra
|
growth hormone releasing hormone receptor a |
| chr1_-_42289704 | 2.14 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr21_-_10446405 | 2.13 |
ENSDART00000167948
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr16_+_50434668 | 2.13 |
ENSDART00000193500
|
IGLON5
|
zgc:110372 |
| chr12_+_16281312 | 2.10 |
ENSDART00000152500
|
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr24_-_11309477 | 2.10 |
ENSDART00000137257
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr10_-_2524917 | 2.10 |
ENSDART00000188642
|
CU856539.1
|
|
| chr2_-_38284648 | 2.10 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr2_-_10188598 | 2.07 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr11_-_37425407 | 2.05 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr9_-_27649406 | 2.05 |
ENSDART00000181270
ENSDART00000187112 |
stxbp5l
|
syntaxin binding protein 5-like |
| chr25_-_210730 | 2.04 |
ENSDART00000187580
|
FP236318.1
|
|
| chr20_+_23173710 | 2.00 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr8_-_44868020 | 2.00 |
ENSDART00000142712
|
cacna1fa
|
calcium channel, voltage-dependent, L type, alpha 1F subunit a |
| chr7_-_27685365 | 2.00 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr15_-_14884332 | 1.99 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr16_-_41439659 | 1.97 |
ENSDART00000191624
|
cpne4a
|
copine IVa |
| chr17_+_6828550 | 1.96 |
ENSDART00000153777
|
si:ch73-242m19.1
|
si:ch73-242m19.1 |
| chr7_+_29167744 | 1.95 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr7_+_15872357 | 1.95 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr21_-_17482465 | 1.94 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr2_+_15203322 | 1.93 |
ENSDART00000144171
|
abca4b
|
ATP-binding cassette, sub-family A (ABC1), member 4b |
| chr6_+_37894220 | 1.93 |
ENSDART00000087311
|
oca2
|
oculocutaneous albinism II |
| chr11_+_6902306 | 1.92 |
ENSDART00000075993
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr20_+_42978499 | 1.92 |
ENSDART00000138793
|
si:ch211-203k16.3
|
si:ch211-203k16.3 |
| chr14_-_34044369 | 1.92 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr13_-_11035420 | 1.91 |
ENSDART00000108709
|
cep170aa
|
centrosomal protein 170Aa |
| chr14_-_46173265 | 1.91 |
ENSDART00000164321
|
zmp:0000000758
|
zmp:0000000758 |
| chr9_+_36314867 | 1.90 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr16_-_27174373 | 1.90 |
ENSDART00000166681
|
frrs1l
|
ferric-chelate reductase 1-like |
| chr4_+_9279784 | 1.88 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr7_+_56098590 | 1.87 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr20_-_30920356 | 1.87 |
ENSDART00000022951
|
kif25
|
kinesin family member 25 |
| chr4_+_5835393 | 1.86 |
ENSDART00000055414
|
pex26
|
peroxisomal biogenesis factor 26 |
| chr1_-_50859053 | 1.86 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr19_+_37701450 | 1.86 |
ENSDART00000087694
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr6_+_52350443 | 1.83 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr5_+_36781732 | 1.82 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr3_-_24980067 | 1.82 |
ENSDART00000048871
|
desi1a
|
desumoylating isopeptidase 1a |
| chr15_-_14625373 | 1.82 |
ENSDART00000171841
|
slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr19_+_9174166 | 1.80 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr16_+_20161805 | 1.80 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr19_-_47323267 | 1.80 |
ENSDART00000190077
|
CU138512.1
|
|
| chr20_-_43775495 | 1.80 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr7_+_854237 | 1.77 |
ENSDART00000181029
|
zgc:175177
|
zgc:175177 |
| chr24_-_23942722 | 1.75 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr1_-_10647307 | 1.75 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr18_+_33171765 | 1.75 |
ENSDART00000064153
|
si:ch73-125k17.2
|
si:ch73-125k17.2 |
| chr7_+_35075847 | 1.74 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr4_+_39742119 | 1.71 |
ENSDART00000176004
|
CR749167.2
|
|
| chr2_+_55429988 | 1.70 |
ENSDART00000166920
|
ap1m1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr9_+_47966167 | 1.69 |
ENSDART00000183345
|
gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr6_-_3573716 | 1.68 |
ENSDART00000026693
|
dmbx1b
|
diencephalon/mesencephalon homeobox 1b |
| chr25_+_26895394 | 1.68 |
ENSDART00000155820
|
si:dkey-42p14.3
|
si:dkey-42p14.3 |
| chr13_-_2520843 | 1.68 |
ENSDART00000187313
ENSDART00000170333 |
cox20
|
COX20 cytochrome c oxidase assembly factor |
| chr10_+_41765944 | 1.67 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr13_-_31622195 | 1.66 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr3_+_46628885 | 1.66 |
ENSDART00000006602
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr8_+_35212233 | 1.66 |
ENSDART00000111021
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr9_+_48219111 | 1.65 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr14_-_31893996 | 1.65 |
ENSDART00000173222
|
gpr101
|
G protein-coupled receptor 101 |
| chr3_+_32594357 | 1.65 |
ENSDART00000151639
|
unc119.2
|
unc-119 lipid binding chaperone B homolog 2 |
| chr19_+_4139065 | 1.64 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr11_+_24001993 | 1.63 |
ENSDART00000168215
|
chia.2
|
chitinase, acidic.2 |
| chr4_-_5018705 | 1.63 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr7_-_69636502 | 1.61 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr1_-_44899287 | 1.61 |
ENSDART00000187522
|
tcirg1a
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3a |
| chr9_+_29548195 | 1.59 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr3_+_34919810 | 1.58 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr23_+_579893 | 1.58 |
ENSDART00000189098
|
BX323461.1
|
|
| chr13_-_14487524 | 1.56 |
ENSDART00000141103
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr5_+_42400777 | 1.54 |
ENSDART00000183114
|
BX548073.8
|
|
| chr22_+_1556948 | 1.54 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr1_-_40911332 | 1.54 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr25_+_5755314 | 1.54 |
ENSDART00000172088
ENSDART00000164208 |
appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr6_+_8315050 | 1.52 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr10_+_29698467 | 1.51 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr11_-_42554290 | 1.51 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr10_-_28835771 | 1.48 |
ENSDART00000192220
ENSDART00000188436 |
alcama
|
activated leukocyte cell adhesion molecule a |
Network of associatons between targets according to the STRING database.
First level regulatory network of prrx1a+prrx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.6 | 10.9 | GO:0045299 | otolith mineralization(GO:0045299) |
| 1.5 | 6.0 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 1.5 | 4.4 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.3 | 5.3 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 1.0 | 49.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.9 | 3.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.9 | 2.7 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.8 | 2.3 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.7 | 3.6 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.7 | 2.8 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.6 | 1.9 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.6 | 3.7 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.6 | 5.6 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.5 | 9.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.5 | 7.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.5 | 1.9 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.4 | 1.8 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.4 | 1.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 1.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.4 | 1.7 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.4 | 2.9 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.4 | 1.2 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.4 | 3.9 | GO:0070836 | membrane raft assembly(GO:0001765) membrane raft organization(GO:0031579) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.4 | 4.2 | GO:0030431 | sleep(GO:0030431) |
| 0.4 | 2.7 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 3.0 | GO:0071632 | optomotor response(GO:0071632) |
| 0.4 | 1.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.3 | 1.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 1.0 | GO:1904377 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.3 | 6.6 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.3 | 3.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.3 | 2.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 2.6 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.3 | 6.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 1.9 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.3 | 4.1 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.3 | 3.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 5.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 4.0 | GO:0036065 | fucosylation(GO:0036065) |
| 0.3 | 1.0 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.2 | 1.2 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 0.7 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.2 | 0.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 2.3 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 3.9 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.2 | 1.3 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 0.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 1.9 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.2 | 2.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 2.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 1.2 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.2 | 3.8 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.2 | 0.8 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 2.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 1.5 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.2 | 2.1 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.2 | 2.2 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.2 | 1.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 11.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.2 | 0.3 | GO:0048320 | axial mesoderm formation(GO:0048320) axial mesodermal cell differentiation(GO:0048321) axial mesodermal cell fate commitment(GO:0048322) axial mesodermal cell fate specification(GO:0048327) |
| 0.2 | 2.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 3.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 3.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.2 | 1.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.6 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.9 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 4.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 2.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 1.4 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 2.3 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.9 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 1.5 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 1.9 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 1.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 4.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 1.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.9 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.9 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 1.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.6 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 2.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 1.0 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 4.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.1 | 2.1 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 3.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 0.9 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 4.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 6.4 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 1.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 2.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 1.0 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 0.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 1.4 | GO:0010257 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 2.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.1 | GO:0060078 | regulation of postsynaptic membrane potential(GO:0060078) |
| 0.1 | 1.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 2.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.6 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.6 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 2.5 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.6 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.6 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 | 3.6 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 1.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.4 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.1 | 1.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 2.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 2.1 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.1 | 3.9 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.8 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 2.0 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 2.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 27.1 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.1 | 1.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.1 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 4.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 2.7 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.5 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 1.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.9 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 1.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.6 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 1.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 4.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 2.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.9 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 1.5 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 1.3 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 2.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.9 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.3 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 3.9 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 1.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.5 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 7.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 2.2 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 2.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.7 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.0 | 0.3 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 1.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 4.6 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.2 | GO:0070265 | necrotic cell death(GO:0070265) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.4 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.8 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 1.9 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 2.5 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.8 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.4 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.4 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.6 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.9 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 1.2 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.0 | 1.5 | GO:0030902 | hindbrain development(GO:0030902) |
| 0.0 | 1.8 | GO:0070646 | protein modification by small protein removal(GO:0070646) |
| 0.0 | 1.4 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.7 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 0.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.7 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.7 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 49.3 | GO:0005861 | troponin complex(GO:0005861) |
| 1.0 | 7.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.8 | 3.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.7 | 2.2 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.6 | 3.6 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.6 | 5.6 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.4 | 6.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 6.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 1.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 11.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.3 | 3.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 1.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 1.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 0.9 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.2 | 4.5 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 1.4 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 5.0 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.6 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 2.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.0 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 8.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 3.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.6 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 2.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 2.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 4.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 4.1 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 2.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 4.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 4.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 0.6 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 2.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 4.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.3 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 3.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 4.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 2.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 10.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 3.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.0 | 0.3 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.3 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.0 | 1.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 9.5 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.0 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.9 | 3.7 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.9 | 2.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.8 | 6.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.8 | 2.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.7 | 2.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.7 | 2.2 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.7 | 3.6 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.7 | 2.0 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.7 | 4.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.6 | 3.9 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.6 | 3.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.6 | 3.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 4.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.5 | 2.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 7.6 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.4 | 5.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 1.3 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.4 | 2.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.4 | 1.5 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.3 | 4.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 4.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 2.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 6.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 4.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 5.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 1.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 2.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 1.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 2.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 1.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 8.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 2.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.2 | 2.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.2 | 2.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 3.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 1.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.9 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 4.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 2.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 5.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 2.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 2.8 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 2.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.7 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 2.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.6 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 1.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 4.8 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 3.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 1.3 | GO:0010851 | cyclase regulator activity(GO:0010851) |
| 0.1 | 0.6 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 1.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.0 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 3.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.4 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 4.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 1.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 2.1 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.7 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 3.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 2.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.6 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.1 | 1.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.8 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 3.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 8.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.9 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 5.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 2.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.5 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0035255 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.6 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 4.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.8 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 13.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 1.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.1 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 20.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 1.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 3.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.8 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 4.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 1.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 3.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 3.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.7 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 4.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 2.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 2.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 1.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 2.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 9.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 2.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 1.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 8.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.4 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 2.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |