Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
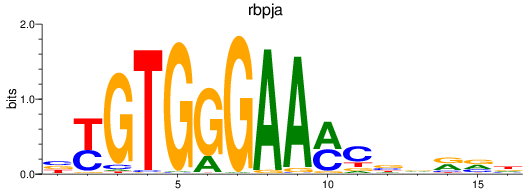
Results for rbpja
Z-value: 0.93
Transcription factors associated with rbpja
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rbpja
|
ENSDARG00000003398 | recombination signal binding protein for immunoglobulin kappa J region a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rbpja | dr11_v1_chr1_+_14454663_14454663 | -0.05 | 6.2e-01 | Click! |
Activity profile of rbpja motif
Sorted Z-values of rbpja motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_4331117 | 12.46 |
ENSDART00000008893
|
ca15a
|
carbonic anhydrase XVa |
| chr22_-_237651 | 9.68 |
ENSDART00000075210
|
zgc:66156
|
zgc:66156 |
| chr1_+_1805294 | 9.31 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr3_-_3209432 | 8.83 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr14_+_51098036 | 8.80 |
ENSDART00000184118
|
CABZ01078594.1
|
|
| chr3_+_26081343 | 8.79 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr17_+_24851951 | 8.18 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr19_-_5332784 | 6.81 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr10_-_20357013 | 6.42 |
ENSDART00000080143
|
sfrp1b
|
secreted frizzled-related protein 1b |
| chr7_-_3429874 | 6.41 |
ENSDART00000132330
|
si:ch211-285c6.1
|
si:ch211-285c6.1 |
| chr6_+_45932276 | 6.11 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr23_+_1730663 | 6.06 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr23_-_27571667 | 6.02 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr9_-_40765868 | 5.76 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr2_-_20923864 | 5.72 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr18_-_46241775 | 5.40 |
ENSDART00000145999
ENSDART00000134244 ENSDART00000185021 ENSDART00000181855 |
prx
|
periaxin |
| chr16_-_45094599 | 5.37 |
ENSDART00000155479
|
si:rp71-77l1.1
|
si:rp71-77l1.1 |
| chr17_+_16046132 | 5.18 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr25_-_18470695 | 4.77 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr20_+_3934516 | 4.72 |
ENSDART00000165732
|
clec11a
|
C-type lectin domain containing 11A |
| chr4_-_20043484 | 4.61 |
ENSDART00000167780
|
zgc:193726
|
zgc:193726 |
| chr21_-_27213166 | 4.53 |
ENSDART00000146959
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr3_-_31254379 | 4.39 |
ENSDART00000189376
|
apnl
|
actinoporin-like protein |
| chr17_+_33433576 | 4.13 |
ENSDART00000077581
|
snap23.2
|
synaptosomal-associated protein 23.2 |
| chr20_-_8110672 | 4.00 |
ENSDART00000113993
|
si:ch211-232i5.1
|
si:ch211-232i5.1 |
| chr10_-_25816558 | 3.93 |
ENSDART00000017240
|
postna
|
periostin, osteoblast specific factor a |
| chr5_-_37900350 | 3.81 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr2_-_30324610 | 3.80 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr23_+_19790962 | 3.79 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr16_+_41974438 | 3.75 |
ENSDART00000102771
|
si:ch211-225p5.8
|
si:ch211-225p5.8 |
| chr20_+_218886 | 3.73 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr15_-_31147301 | 3.63 |
ENSDART00000157145
ENSDART00000155473 ENSDART00000048103 |
ksr1b
|
kinase suppressor of ras 1b |
| chr9_-_56399699 | 3.61 |
ENSDART00000170281
|
rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chr25_+_245018 | 3.60 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr16_-_44945224 | 3.60 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr3_-_53092509 | 3.59 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr18_-_5209258 | 3.53 |
ENSDART00000183109
|
CABZ01080601.1
|
|
| chr2_-_30324297 | 3.44 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr4_+_55778679 | 3.43 |
ENSDART00000183009
|
CT583728.19
|
|
| chr4_+_55794876 | 3.43 |
ENSDART00000189043
|
CT583728.17
|
|
| chr4_+_55810436 | 3.43 |
ENSDART00000182875
|
CT583728.16
|
|
| chr4_-_68568233 | 3.42 |
ENSDART00000184284
|
BX548011.1
|
|
| chr3_+_36646054 | 3.40 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr19_+_38422059 | 3.34 |
ENSDART00000035093
|
col9a2
|
procollagen, type IX, alpha 2 |
| chr14_+_26229056 | 3.31 |
ENSDART00000179045
|
LO018208.1
|
|
| chr16_+_2905150 | 3.21 |
ENSDART00000109980
|
lars2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr16_+_26449615 | 3.18 |
ENSDART00000039746
|
epb41b
|
erythrocyte membrane protein band 4.1b |
| chr21_-_22720381 | 3.12 |
ENSDART00000130789
|
c1qc
|
complement component 1, q subcomponent, C chain |
| chr3_+_4213864 | 3.12 |
ENSDART00000059613
|
si:dkeyp-52c3.5
|
si:dkeyp-52c3.5 |
| chr2_-_32237916 | 3.07 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr25_+_20272145 | 3.01 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr13_-_34781984 | 3.01 |
ENSDART00000172138
|
ism1
|
isthmin 1 |
| chr7_+_49681040 | 3.00 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr3_+_2669813 | 3.00 |
ENSDART00000014205
|
CR388047.1
|
|
| chr4_-_74028161 | 3.00 |
ENSDART00000174062
|
LO018205.1
|
|
| chr23_-_32092443 | 2.96 |
ENSDART00000133688
|
letmd1
|
LETM1 domain containing 1 |
| chr10_-_342564 | 2.95 |
ENSDART00000157633
|
vaspa
|
vasodilator stimulated phosphoprotein a |
| chr17_-_2573021 | 2.94 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr16_-_24605969 | 2.94 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr20_+_23501535 | 2.90 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr5_-_6567464 | 2.90 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr12_-_30548244 | 2.87 |
ENSDART00000193616
|
zgc:158404
|
zgc:158404 |
| chr25_+_34915762 | 2.86 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr6_-_9695294 | 2.83 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr23_-_5783421 | 2.79 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr19_-_18127808 | 2.76 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr20_+_53368611 | 2.73 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr25_+_245438 | 2.71 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr19_+_42609132 | 2.69 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr19_+_48018802 | 2.65 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr22_-_506522 | 2.57 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr20_-_39596338 | 2.41 |
ENSDART00000023531
|
hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr1_-_54100988 | 2.39 |
ENSDART00000192662
|
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr19_+_43341424 | 2.36 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr18_-_18937485 | 2.35 |
ENSDART00000139015
|
si:dkey-73n10.1
|
si:dkey-73n10.1 |
| chr11_+_13159988 | 2.30 |
ENSDART00000064176
|
mob3c
|
MOB kinase activator 3C |
| chr5_+_36850650 | 2.26 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr7_+_2236317 | 2.25 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr10_+_38512270 | 2.18 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr20_-_49657134 | 2.17 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr2_+_8112449 | 2.17 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr20_+_39283849 | 2.13 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr19_-_18127629 | 2.13 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr19_-_12648122 | 2.12 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr1_+_52690448 | 2.12 |
ENSDART00000150326
|
osbp
|
oxysterol binding protein |
| chr4_+_21741228 | 2.11 |
ENSDART00000112035
ENSDART00000127664 |
myf5
|
myogenic factor 5 |
| chr24_+_7884880 | 2.10 |
ENSDART00000139467
|
bmp6
|
bone morphogenetic protein 6 |
| chr19_-_42462491 | 2.08 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| chr10_-_44306636 | 2.08 |
ENSDART00000191068
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr4_-_76183357 | 2.08 |
ENSDART00000181362
ENSDART00000150331 |
si:ch211-106j21.4
zgc:110171
|
si:ch211-106j21.4 zgc:110171 |
| chr16_-_17300030 | 2.07 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr10_-_44306399 | 2.07 |
ENSDART00000180042
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr3_+_3641429 | 2.03 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr18_-_21746421 | 2.02 |
ENSDART00000188809
|
pskh1
|
protein serine kinase H1 |
| chr5_-_32338866 | 2.01 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr11_-_22361306 | 2.00 |
ENSDART00000180688
ENSDART00000182200 ENSDART00000006580 |
tfeb
|
transcription factor EB |
| chr16_-_25680666 | 1.96 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr16_-_30885838 | 1.96 |
ENSDART00000131356
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr22_-_164944 | 1.95 |
ENSDART00000145379
|
fblim1
|
filamin binding LIM protein 1 |
| chr12_-_46959990 | 1.91 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr10_-_38468847 | 1.90 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr19_+_3842891 | 1.86 |
ENSDART00000159043
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr8_-_46386024 | 1.79 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr24_-_9989634 | 1.74 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr21_+_45268112 | 1.70 |
ENSDART00000157136
|
tcf7
|
transcription factor 7 |
| chr21_+_20549395 | 1.68 |
ENSDART00000181633
|
efna5a
|
ephrin-A5a |
| chr5_-_47727819 | 1.64 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr1_-_51720633 | 1.64 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr12_+_9542124 | 1.64 |
ENSDART00000127952
|
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr4_-_39484284 | 1.62 |
ENSDART00000129216
|
si:dkey-261o4.6
|
si:dkey-261o4.6 |
| chr16_+_9762261 | 1.61 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr5_+_9246458 | 1.60 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr3_+_17616201 | 1.58 |
ENSDART00000156775
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr11_-_16394971 | 1.58 |
ENSDART00000180981
ENSDART00000179925 |
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr13_+_674351 | 1.55 |
ENSDART00000111328
|
cutc
|
cutC copper transporter homolog (E. coli) |
| chr13_-_49819027 | 1.52 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr16_-_32233463 | 1.52 |
ENSDART00000102016
|
calhm6
|
calcium homeostasis modulator family member 6 |
| chr12_+_7399403 | 1.50 |
ENSDART00000103526
|
bicc1b
|
BicC family RNA binding protein 1b |
| chr17_+_5061135 | 1.50 |
ENSDART00000064313
|
cdc5l
|
CDC5 cell division cycle 5-like (S. pombe) |
| chr14_+_32926385 | 1.47 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr24_-_21090447 | 1.45 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr24_-_37877978 | 1.45 |
ENSDART00000128574
|
tmem204
|
transmembrane protein 204 |
| chr18_-_50676567 | 1.43 |
ENSDART00000172264
|
ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr23_+_17839187 | 1.40 |
ENSDART00000104647
|
prim1
|
DNA primase subunit 1 |
| chr24_+_35933869 | 1.40 |
ENSDART00000173322
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr20_+_14789148 | 1.39 |
ENSDART00000164761
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr2_-_50053006 | 1.39 |
ENSDART00000083654
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr5_-_17876709 | 1.38 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr15_-_28908027 | 1.37 |
ENSDART00000182790
ENSDART00000192461 |
eml2
|
echinoderm microtubule associated protein like 2 |
| chr1_-_23370395 | 1.36 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr3_-_3428938 | 1.33 |
ENSDART00000179811
ENSDART00000115282 ENSDART00000192263 ENSDART00000046454 |
A2ML1 (1 of many)
|
zgc:171445 |
| chr16_-_30901159 | 1.32 |
ENSDART00000180313
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr20_-_3911546 | 1.26 |
ENSDART00000169787
|
cnksr3
|
cnksr family member 3 |
| chr14_+_13454840 | 1.25 |
ENSDART00000161854
|
pls3
|
plastin 3 (T isoform) |
| chr7_+_65564163 | 1.25 |
ENSDART00000082679
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr8_+_36554816 | 1.25 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr7_-_5125799 | 1.23 |
ENSDART00000173390
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr12_-_35054354 | 1.22 |
ENSDART00000075351
|
zgc:112285
|
zgc:112285 |
| chr10_+_31951338 | 1.22 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr14_-_4321874 | 1.21 |
ENSDART00000042672
|
guf1
|
GUF1 homolog, GTPase |
| chr19_+_48018464 | 1.19 |
ENSDART00000172307
ENSDART00000163848 |
UBE2M
|
si:ch1073-205c8.3 |
| chr15_-_28907709 | 1.16 |
ENSDART00000017268
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr4_-_74367912 | 1.14 |
ENSDART00000174199
ENSDART00000165257 |
ptprb
|
protein tyrosine phosphatase, receptor type, b |
| chr25_+_1549838 | 1.14 |
ENSDART00000115001
|
avpr1aa
|
arginine vasopressin receptor 1Aa |
| chr12_+_25223843 | 1.13 |
ENSDART00000077180
ENSDART00000127454 ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr10_+_573667 | 1.09 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr24_-_37877554 | 1.08 |
ENSDART00000132219
|
tmem204
|
transmembrane protein 204 |
| chr2_+_3201345 | 1.07 |
ENSDART00000130349
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr21_-_37194365 | 1.06 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr3_-_3439150 | 1.05 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr7_+_3558030 | 1.00 |
ENSDART00000158322
|
si:dkey-192d15.1
|
si:dkey-192d15.1 |
| chr2_-_59247811 | 1.00 |
ENSDART00000141384
|
ftr32
|
finTRIM family, member 32 |
| chr6_-_36552844 | 0.99 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr7_+_54475134 | 0.96 |
ENSDART00000164063
|
zgc:153993
|
zgc:153993 |
| chr17_+_11372531 | 0.96 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr24_-_25184553 | 0.96 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr24_+_39027481 | 0.95 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr4_+_45441669 | 0.94 |
ENSDART00000150745
|
si:ch211-162i8.7
|
si:ch211-162i8.7 |
| chr4_+_74943111 | 0.94 |
ENSDART00000004739
|
nup50
|
nucleoporin 50 |
| chr10_-_39091894 | 0.92 |
ENSDART00000184505
|
igsf5a
|
immunoglobulin superfamily, member 5a |
| chr22_-_26274177 | 0.91 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr5_-_57641257 | 0.91 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr3_+_31058464 | 0.87 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr20_+_14789305 | 0.87 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr1_-_46981134 | 0.84 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr20_-_33961697 | 0.82 |
ENSDART00000061765
|
selp
|
selectin P |
| chr10_-_10864331 | 0.77 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr18_-_18587745 | 0.77 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr18_-_46369516 | 0.75 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr20_-_54508650 | 0.71 |
ENSDART00000147642
|
CABZ01118770.1
|
|
| chr13_+_21919786 | 0.70 |
ENSDART00000182440
|
ndst2a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2a |
| chr19_+_41520892 | 0.68 |
ENSDART00000182218
ENSDART00000115123 |
crtc2
|
CREB regulated transcription coactivator 2 |
| chr4_-_65210616 | 0.67 |
ENSDART00000183988
ENSDART00000186858 ENSDART00000185740 |
si:dkey-14o6.1
|
si:dkey-14o6.1 |
| chr2_-_898899 | 0.67 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr23_-_24047054 | 0.66 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr18_-_17075098 | 0.66 |
ENSDART00000042496
ENSDART00000192284 ENSDART00000180307 |
tango6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr3_+_27722355 | 0.62 |
ENSDART00000132761
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr2_-_55317035 | 0.59 |
ENSDART00000169382
ENSDART00000097874 |
tpm4b
|
tropomyosin 4b |
| chr21_-_22831388 | 0.57 |
ENSDART00000151040
|
angptl5
|
angiopoietin-like 5 |
| chr6_-_59388625 | 0.57 |
ENSDART00000111992
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr25_+_2263857 | 0.57 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr17_-_6508406 | 0.53 |
ENSDART00000002778
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr10_-_10863936 | 0.53 |
ENSDART00000180568
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr25_+_16646113 | 0.50 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr19_+_10536093 | 0.50 |
ENSDART00000138706
|
si:dkey-211g8.1
|
si:dkey-211g8.1 |
| chr22_-_3255341 | 0.50 |
ENSDART00000114232
|
gpr35.1
|
G protein-coupled receptor 35, tandem duplicate 1 |
| chr7_+_15329819 | 0.49 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr25_+_34915576 | 0.48 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr7_+_39054478 | 0.47 |
ENSDART00000173825
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr6_+_58522738 | 0.46 |
ENSDART00000157327
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr2_-_38287987 | 0.43 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr2_-_41518340 | 0.42 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr8_+_16726386 | 0.40 |
ENSDART00000144621
|
smim15
|
small integral membrane protein 15 |
| chr7_+_61184104 | 0.39 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr7_+_19495379 | 0.38 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr13_-_50546634 | 0.37 |
ENSDART00000192127
|
CU570781.2
|
|
| chr10_+_39091353 | 0.37 |
ENSDART00000125986
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr19_-_24218942 | 0.37 |
ENSDART00000189198
|
BX547993.2
|
|
| chr14_+_109016 | 0.37 |
ENSDART00000158405
|
gpc2
|
glypican 2 |
| chr7_+_24496894 | 0.37 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of rbpja
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.8 | GO:0090076 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 1.4 | 5.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.1 | 12.5 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 1.0 | 4.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.8 | 2.4 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.8 | 5.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.7 | 4.4 | GO:0051818 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.7 | 5.8 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.7 | 3.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.7 | 2.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.7 | 3.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.5 | 6.0 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.5 | 3.7 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.5 | 9.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.4 | 1.7 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.4 | 1.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.4 | 1.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 2.4 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.3 | 2.6 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.3 | 6.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 1.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 2.0 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.2 | 1.9 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 3.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 1.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 3.7 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.2 | 4.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 3.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 3.0 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.2 | 3.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.2 | 1.6 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.1 | 4.1 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.8 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 2.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 6.4 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 1.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 3.0 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 1.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 2.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 2.2 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 2.0 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 2.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.7 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.0 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.1 | 3.1 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 2.9 | GO:2000649 | regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 2.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 2.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.4 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 2.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 3.1 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 3.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.5 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.2 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 4.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 4.7 | GO:0001503 | ossification(GO:0001503) |
| 0.0 | 9.4 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.4 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 1.2 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.6 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.4 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 1.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 2.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 3.0 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 1.7 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 1.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.4 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 7.1 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 3.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.4 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 4.1 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.2 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 3.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.7 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.9 | GO:0006400 | tRNA modification(GO:0006400) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.8 | GO:0031673 | H zone(GO:0031673) |
| 1.5 | 4.4 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.4 | 7.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.7 | 3.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.6 | 1.7 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 6.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.5 | 2.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.5 | 1.4 | GO:1990077 | primosome complex(GO:1990077) |
| 0.4 | 1.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 2.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 2.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 3.7 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 1.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 3.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 3.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 3.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 2.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 2.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 9.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 6.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 12.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 4.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 8.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 5.2 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 2.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.7 | GO:0030425 | dendrite(GO:0030425) somatodendritic compartment(GO:0036477) |
| 0.0 | 0.4 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.9 | 9.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.7 | 3.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 6.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.5 | 8.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.5 | 2.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.5 | 1.9 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.5 | 3.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.4 | 12.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 3.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 2.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 6.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 3.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 2.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 3.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 2.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 1.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 2.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.2 | 4.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 6.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 1.9 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 1.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 2.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 0.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 4.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.8 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 3.0 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.6 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 2.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 2.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 4.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 2.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 2.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 4.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 2.0 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 3.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 5.8 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 1.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 5.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 4.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 9.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 4.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 12.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 2.9 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.5 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 8.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 3.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 3.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 4.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.5 | 5.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.5 | 8.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 2.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 3.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 5.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 2.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 1.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 6.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.7 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 2.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 5.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.6 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |