Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
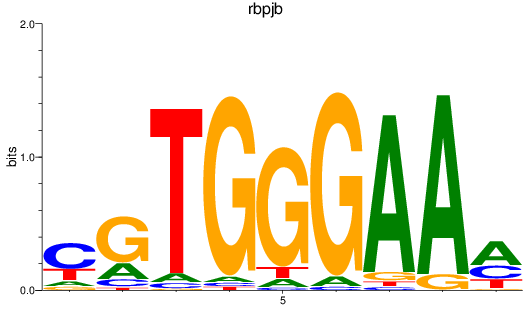
Results for rbpjb
Z-value: 0.82
Transcription factors associated with rbpjb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rbpjb
|
ENSDARG00000052091 | recombination signal binding protein for immunoglobulin kappa J region b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rbpjb | dr11_v1_chr7_+_62080456_62080456 | 0.07 | 5.3e-01 | Click! |
Activity profile of rbpjb motif
Sorted Z-values of rbpjb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_31891110 | 11.68 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr17_-_25326685 | 8.06 |
ENSDART00000082327
|
pabpc4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr17_-_25326296 | 7.15 |
ENSDART00000168822
|
pabpc4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr10_+_22782522 | 6.63 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr25_-_23526058 | 5.09 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr20_-_39596338 | 4.88 |
ENSDART00000023531
|
hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr11_-_11518469 | 4.79 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr3_-_36839115 | 4.25 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr22_+_16308806 | 3.96 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr22_+_16308450 | 3.96 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr2_-_898899 | 3.59 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr19_+_42609132 | 3.57 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr25_+_6122823 | 3.55 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr19_-_25113660 | 3.39 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr16_-_42523744 | 3.32 |
ENSDART00000017185
|
tbx20
|
T-box 20 |
| chr3_+_41917499 | 3.21 |
ENSDART00000028673
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr15_-_37846047 | 3.11 |
ENSDART00000184837
|
hsc70
|
heat shock cognate 70 |
| chr2_-_44746723 | 3.09 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium chain family member 3 |
| chr4_-_9722568 | 3.07 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr6_-_36552844 | 2.95 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr5_-_69312533 | 2.84 |
ENSDART00000082614
ENSDART00000183098 |
smtnb
|
smoothelin b |
| chr23_-_23401305 | 2.75 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr21_-_27213166 | 2.74 |
ENSDART00000146959
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr24_-_6375774 | 2.69 |
ENSDART00000013294
|
anxa13
|
annexin A13 |
| chr1_-_45633955 | 2.62 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr5_+_23151169 | 2.46 |
ENSDART00000125638
|
tbx5b
|
T-box 5b |
| chr20_+_39283849 | 2.45 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr9_+_38684871 | 2.35 |
ENSDART00000147938
ENSDART00000004462 |
heg1
|
heart development protein with EGF-like domains 1 |
| chr22_+_30335936 | 2.33 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr24_-_37877743 | 2.14 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr7_+_27834130 | 2.04 |
ENSDART00000052656
|
rras2
|
RAS related 2 |
| chr6_+_45932276 | 2.03 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr5_-_47727819 | 2.03 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr19_-_31802296 | 1.93 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr13_+_30804367 | 1.89 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr13_+_33246150 | 1.89 |
ENSDART00000136356
ENSDART00000132834 ENSDART00000144379 ENSDART00000131985 ENSDART00000148368 ENSDART00000035940 ENSDART00000141912 |
ndufaf1
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr10_-_10863936 | 1.86 |
ENSDART00000180568
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr10_-_10864331 | 1.83 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr4_-_74367912 | 1.82 |
ENSDART00000174199
ENSDART00000165257 |
ptprb
|
protein tyrosine phosphatase, receptor type, b |
| chr14_+_4151379 | 1.80 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr20_+_36234335 | 1.77 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr10_+_9195190 | 1.71 |
ENSDART00000136364
|
antxr2b
|
anthrax toxin receptor 2b |
| chr6_-_14292307 | 1.68 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr7_+_38445938 | 1.55 |
ENSDART00000173467
|
cep89
|
centrosomal protein 89 |
| chr16_+_26449615 | 1.51 |
ENSDART00000039746
|
epb41b
|
erythrocyte membrane protein band 4.1b |
| chr7_-_15185811 | 1.51 |
ENSDART00000031049
|
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr24_-_37877978 | 1.43 |
ENSDART00000128574
|
tmem204
|
transmembrane protein 204 |
| chr11_+_13159988 | 1.42 |
ENSDART00000064176
|
mob3c
|
MOB kinase activator 3C |
| chr10_-_42297889 | 1.37 |
ENSDART00000099262
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr23_-_31372639 | 1.35 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr15_+_19783352 | 1.34 |
ENSDART00000178784
ENSDART00000092560 |
zpld1b
|
zona pellucida-like domain containing 1b |
| chr16_-_17300030 | 1.34 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr18_+_22220656 | 1.30 |
ENSDART00000191862
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr3_+_27722355 | 1.26 |
ENSDART00000132761
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr16_+_36671064 | 1.25 |
ENSDART00000109703
|
col6a4a
|
collagen, type VI, alpha 4a |
| chr17_+_30546579 | 1.24 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr13_+_33655404 | 1.18 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr13_-_36579086 | 1.16 |
ENSDART00000146671
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr17_+_24851951 | 1.14 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr8_-_18613948 | 1.13 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr22_+_12431608 | 1.12 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr19_+_48018464 | 1.10 |
ENSDART00000172307
ENSDART00000163848 |
UBE2M
|
si:ch1073-205c8.3 |
| chr1_-_51720633 | 1.08 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr8_-_29706882 | 1.08 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr8_+_48491387 | 1.07 |
ENSDART00000086053
|
PRDM16
|
si:ch211-263k4.2 |
| chr5_-_56513825 | 1.07 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr14_-_33083539 | 1.07 |
ENSDART00000160173
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr22_-_30881738 | 1.06 |
ENSDART00000059965
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr13_-_25305179 | 1.04 |
ENSDART00000110064
|
plaua
|
plasminogen activator, urokinase a |
| chr14_+_23076207 | 1.04 |
ENSDART00000161628
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr13_-_24874950 | 1.03 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr19_+_48018802 | 1.01 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr16_+_46492994 | 1.01 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr5_+_46424437 | 1.01 |
ENSDART00000186511
|
vcana
|
versican a |
| chr23_-_8733092 | 1.00 |
ENSDART00000014222
|
tcea2
|
transcription elongation factor A (SII), 2 |
| chr15_-_41714329 | 0.99 |
ENSDART00000154113
|
CR853294.1
|
|
| chr21_-_5799122 | 0.98 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr24_-_37877554 | 0.98 |
ENSDART00000132219
|
tmem204
|
transmembrane protein 204 |
| chr3_-_34547000 | 0.98 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr9_-_23807032 | 0.96 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr13_-_33654931 | 0.95 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr13_-_36798204 | 0.91 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr13_-_33009734 | 0.91 |
ENSDART00000134140
|
rbm25a
|
RNA binding motif protein 25a |
| chr2_+_2110271 | 0.87 |
ENSDART00000148460
|
dspa
|
desmoplakin a |
| chr15_+_20530649 | 0.86 |
ENSDART00000186312
|
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr14_+_38786298 | 0.86 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr3_-_54992934 | 0.86 |
ENSDART00000053095
ENSDART00000145766 |
rhbdf1a
|
rhomboid 5 homolog 1a (Drosophila) |
| chr24_-_20658446 | 0.85 |
ENSDART00000127923
|
nktr
|
natural killer cell triggering receptor |
| chr4_+_4849789 | 0.84 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr5_+_9428876 | 0.84 |
ENSDART00000081791
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr9_-_3671911 | 0.84 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr12_-_19862912 | 0.82 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr19_-_7540821 | 0.78 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr17_+_49081828 | 0.77 |
ENSDART00000156492
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr5_-_36597612 | 0.76 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr14_+_8475007 | 0.75 |
ENSDART00000148210
|
dnajc4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr24_-_24796583 | 0.75 |
ENSDART00000144791
ENSDART00000146570 |
pde7a
|
phosphodiesterase 7A |
| chr5_-_66028714 | 0.75 |
ENSDART00000022625
ENSDART00000164228 |
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr24_+_28953089 | 0.74 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr15_-_47468085 | 0.71 |
ENSDART00000164438
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr19_-_32641725 | 0.70 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr8_-_11067079 | 0.68 |
ENSDART00000181986
|
dennd2c
|
DENN/MADD domain containing 2C |
| chr9_+_41218967 | 0.67 |
ENSDART00000000280
ENSDART00000145674 |
stat1b
|
signal transducer and activator of transcription 1b |
| chr19_-_12648408 | 0.64 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr3_+_21669545 | 0.63 |
ENSDART00000156527
|
crhr1
|
corticotropin releasing hormone receptor 1 |
| chr5_+_15202495 | 0.63 |
ENSDART00000144915
|
tbx1
|
T-box 1 |
| chr10_-_4375190 | 0.61 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr12_+_35587971 | 0.61 |
ENSDART00000166072
ENSDART00000181861 ENSDART00000158043 |
chmp6b
|
charged multivesicular body protein 6b |
| chr18_+_26829362 | 0.61 |
ENSDART00000132728
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr13_-_27916439 | 0.60 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr13_-_31389661 | 0.60 |
ENSDART00000134630
|
zdhhc16a
|
zinc finger, DHHC-type containing 16a |
| chr12_+_35067863 | 0.58 |
ENSDART00000152849
|
syt15
|
synaptotagmin XV |
| chr22_-_11729350 | 0.57 |
ENSDART00000105813
|
krt222
|
keratin 222 |
| chr18_+_26829086 | 0.57 |
ENSDART00000098356
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr9_+_19623363 | 0.57 |
ENSDART00000142471
ENSDART00000147662 ENSDART00000136053 |
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr25_+_10793478 | 0.56 |
ENSDART00000058339
ENSDART00000134923 |
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr13_-_3516473 | 0.54 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr19_+_3842891 | 0.54 |
ENSDART00000159043
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr11_-_29563437 | 0.53 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr20_+_45620076 | 0.52 |
ENSDART00000113806
|
hnrnpa0l
|
heterogeneous nuclear ribonucleoprotein A0, like |
| chr24_-_26310854 | 0.52 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr11_-_27962757 | 0.51 |
ENSDART00000147386
|
ece1
|
endothelin converting enzyme 1 |
| chr18_-_16953978 | 0.51 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr21_-_4849029 | 0.51 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr5_+_9348284 | 0.51 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr19_-_11015238 | 0.51 |
ENSDART00000010997
|
tpm3
|
tropomyosin 3 |
| chr8_+_23009662 | 0.49 |
ENSDART00000030885
|
uckl1a
|
uridine-cytidine kinase 1-like 1a |
| chr3_+_46315016 | 0.49 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr9_-_22355391 | 0.48 |
ENSDART00000009115
|
crygm3
|
crystallin, gamma M3 |
| chr24_-_19718077 | 0.47 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr20_-_32045057 | 0.46 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr11_+_34921492 | 0.46 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr18_-_5103931 | 0.45 |
ENSDART00000188091
|
pdcd10a
|
programmed cell death 10a |
| chr7_-_71837213 | 0.45 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr21_-_38852860 | 0.45 |
ENSDART00000166101
|
tlr22
|
toll-like receptor 22 |
| chr21_+_34976600 | 0.43 |
ENSDART00000191672
ENSDART00000139635 |
rbm11
|
RNA binding motif protein 11 |
| chr5_-_57361594 | 0.43 |
ENSDART00000112793
|
cdc26
|
cell division cycle 26 homolog |
| chr3_+_49021079 | 0.40 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr16_-_13881139 | 0.39 |
ENSDART00000138904
ENSDART00000090221 ENSDART00000135099 |
cdc42ep5
|
CDC42 effector protein (Rho GTPase binding) 5 |
| chr12_+_38563073 | 0.39 |
ENSDART00000009172
|
ttyh2
|
tweety family member 2 |
| chr3_-_29869120 | 0.38 |
ENSDART00000138327
|
rpl3
|
ribosomal protein L3 |
| chr25_+_35304903 | 0.38 |
ENSDART00000034313
ENSDART00000187659 |
gas2a
|
growth arrest-specific 2a |
| chr23_-_36003282 | 0.38 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr2_+_3696038 | 0.37 |
ENSDART00000150499
ENSDART00000136906 ENSDART00000164152 ENSDART00000128514 ENSDART00000108964 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr5_-_23675222 | 0.37 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr21_-_43485351 | 0.36 |
ENSDART00000151597
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr20_-_54508650 | 0.36 |
ENSDART00000147642
|
CABZ01118770.1
|
|
| chr23_-_36003441 | 0.36 |
ENSDART00000164699
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr5_-_66028371 | 0.36 |
ENSDART00000183012
|
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr8_+_21159122 | 0.35 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr18_-_6943577 | 0.33 |
ENSDART00000132399
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr9_-_7089303 | 0.32 |
ENSDART00000146609
|
coa5
|
cytochrome C oxidase assembly factor 5 |
| chr20_-_26536581 | 0.31 |
ENSDART00000181810
|
stx11b.2
|
syntaxin 11b, tandem duplicate 2 |
| chr5_+_44944778 | 0.30 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr21_+_20549395 | 0.29 |
ENSDART00000181633
|
efna5a
|
ephrin-A5a |
| chr2_+_55916911 | 0.29 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr8_-_41273461 | 0.28 |
ENSDART00000190598
|
rnf10
|
ring finger protein 10 |
| chr2_+_31957554 | 0.26 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr16_-_21047872 | 0.26 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr8_-_41273768 | 0.26 |
ENSDART00000108518
|
rnf10
|
ring finger protein 10 |
| chr2_-_32769759 | 0.25 |
ENSDART00000178951
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr7_-_1918971 | 0.25 |
ENSDART00000181759
|
CABZ01040626.1
|
|
| chr18_-_11184584 | 0.24 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr18_-_37241080 | 0.24 |
ENSDART00000126421
ENSDART00000078064 |
six9
|
SIX homeobox 9 |
| chr5_+_25736979 | 0.23 |
ENSDART00000175959
|
abhd17b
|
abhydrolase domain containing 17B |
| chr7_+_19424857 | 0.23 |
ENSDART00000173674
|
si:ch211-212k18.6
|
si:ch211-212k18.6 |
| chr6_-_49861506 | 0.23 |
ENSDART00000121809
|
gnas
|
GNAS complex locus |
| chr15_-_31147301 | 0.23 |
ENSDART00000157145
ENSDART00000155473 ENSDART00000048103 |
ksr1b
|
kinase suppressor of ras 1b |
| chr17_+_35362851 | 0.23 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr21_-_40562705 | 0.22 |
ENSDART00000158289
ENSDART00000171997 |
taok1b
|
TAO kinase 1b |
| chr10_+_40624702 | 0.22 |
ENSDART00000109129
|
zgc:172131
|
zgc:172131 |
| chr19_-_12648122 | 0.21 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr23_-_21471022 | 0.21 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr7_+_36041509 | 0.20 |
ENSDART00000162850
|
irx3a
|
iroquois homeobox 3a |
| chr8_+_14915332 | 0.20 |
ENSDART00000164385
|
abl2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr17_-_15777350 | 0.18 |
ENSDART00000155711
|
ankrd6a
|
ankyrin repeat domain 6a |
| chr15_+_5339056 | 0.18 |
ENSDART00000174319
|
CABZ01018874.2
|
|
| chr23_-_31346319 | 0.17 |
ENSDART00000008401
ENSDART00000138106 |
phip
|
pleckstrin homology domain interacting protein |
| chr6_-_25384792 | 0.17 |
ENSDART00000169030
|
PKN2 (1 of many)
|
zgc:153916 |
| chr9_-_14084044 | 0.16 |
ENSDART00000141571
|
fer1l6
|
fer-1-like family member 6 |
| chr2_+_1881334 | 0.16 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr24_+_31374324 | 0.16 |
ENSDART00000172335
ENSDART00000163162 |
cpne3
|
copine III |
| chr3_+_31058464 | 0.16 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr18_-_21177674 | 0.15 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
| chr3_+_37574885 | 0.15 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr22_+_22302614 | 0.15 |
ENSDART00000049434
|
scamp4
|
secretory carrier membrane protein 4 |
| chr19_+_24896409 | 0.15 |
ENSDART00000049840
|
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr1_+_1941031 | 0.15 |
ENSDART00000110331
|
PTGFRN
|
si:ch211-132g1.7 |
| chr6_+_72040 | 0.14 |
ENSDART00000122957
|
crygm4
|
crystallin, gamma M4 |
| chr20_+_5065268 | 0.13 |
ENSDART00000053883
|
si:dkey-60a16.1
|
si:dkey-60a16.1 |
| chr8_+_48848200 | 0.13 |
ENSDART00000130673
|
tp73
|
tumor protein p73 |
| chr23_-_21446985 | 0.13 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr3_-_41995321 | 0.12 |
ENSDART00000192277
|
ttyh3a
|
tweety family member 3a |
| chr12_-_6905375 | 0.12 |
ENSDART00000152322
|
pcdh15b
|
protocadherin-related 15b |
| chr4_+_62598975 | 0.12 |
ENSDART00000163548
|
si:ch211-79g12.2
|
si:ch211-79g12.2 |
| chr2_+_8112449 | 0.12 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr6_-_12172424 | 0.12 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr21_+_43485512 | 0.11 |
ENSDART00000026666
|
atg4a
|
autophagy related 4A, cysteine peptidase |
| chr5_-_33287691 | 0.11 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr2_+_35728033 | 0.11 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rbpjb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.7 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 1.6 | 4.9 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.8 | 3.3 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.8 | 2.5 | GO:0003218 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.7 | 3.5 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.6 | 1.9 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.6 | 3.7 | GO:0055016 | hypochord development(GO:0055016) |
| 0.5 | 1.4 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.4 | 5.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.4 | 1.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.4 | 1.1 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.3 | 1.4 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.3 | 1.0 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.3 | 3.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.3 | 7.9 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.2 | 1.2 | GO:0090280 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) positive regulation of calcium ion import(GO:0090280) |
| 0.2 | 0.6 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.2 | 3.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.2 | 1.0 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 0.6 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.2 | 1.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 1.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 3.6 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 2.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.5 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 2.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.7 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 4.3 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.6 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 1.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.9 | GO:1904950 | negative regulation of protein transport(GO:0051224) negative regulation of establishment of protein localization(GO:1904950) |
| 0.1 | 3.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.1 | 1.2 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 1.1 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.5 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 1.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.5 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 2.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.7 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.5 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 3.4 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.5 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.4 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 5.1 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 1.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.9 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 3.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.5 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 3.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.6 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.2 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 2.3 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.5 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 1.0 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.9 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.5 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 3.4 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.8 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.4 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 7.9 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 1.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.5 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 3.6 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 2.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 4.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 7.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 15.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 1.1 | 11.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.8 | 3.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 1.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 1.2 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.2 | 4.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 3.1 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.2 | 3.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 0.7 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 1.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 1.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 2.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 3.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 7.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 2.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 2.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 3.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 5.1 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 3.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 4.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 3.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 11.3 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 2.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 2.2 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 6.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 4.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |