Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
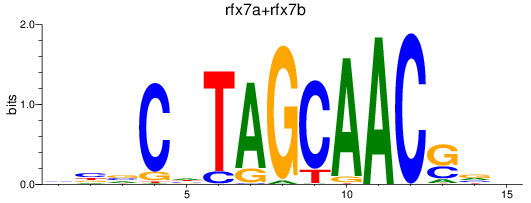
Results for rfx7a+rfx7b
Z-value: 1.00
Transcription factors associated with rfx7a+rfx7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx7a
|
ENSDARG00000077237 | regulatory factor X7a |
|
rfx7b
|
ENSDARG00000103691 | regulatory factor X7b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rfx7a | dr11_v1_chr7_+_34506937_34506937 | 0.47 | 1.4e-06 | Click! |
| rfx7b | dr11_v1_chr25_+_388258_388258 | 0.27 | 8.2e-03 | Click! |
Activity profile of rfx7a+rfx7b motif
Sorted Z-values of rfx7a+rfx7b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_47193564 | 16.24 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr19_+_21266008 | 9.70 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr15_+_22267847 | 9.40 |
ENSDART00000110665
|
spa17
|
sperm autoantigenic protein 17 |
| chr1_+_8662530 | 8.78 |
ENSDART00000054989
|
fscn1b
|
fascin actin-bundling protein 1b |
| chr19_+_2279051 | 8.64 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr25_+_34407740 | 8.53 |
ENSDART00000012677
|
OTUD7A
|
si:dkey-37f18.2 |
| chr19_+_2275019 | 8.51 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr6_+_12853655 | 8.31 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr6_-_33023745 | 7.74 |
ENSDART00000156211
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr20_-_29475172 | 7.19 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr20_-_29474859 | 7.10 |
ENSDART00000152906
ENSDART00000045249 |
scg5
|
secretogranin V |
| chr1_+_604127 | 7.09 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr22_+_3223489 | 7.06 |
ENSDART00000082011
|
lim2.2
|
lens intrinsic membrane protein 2.2 |
| chr14_+_6954579 | 6.84 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr19_-_47526737 | 6.82 |
ENSDART00000186636
|
scg5
|
secretogranin V |
| chr24_-_37472727 | 6.71 |
ENSDART00000134152
|
cluap1
|
clusterin associated protein 1 |
| chr18_+_28102620 | 6.71 |
ENSDART00000132342
|
kiaa1549lb
|
KIAA1549-like b |
| chr17_+_23937262 | 6.65 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr23_+_6272638 | 6.63 |
ENSDART00000190366
|
syt2a
|
synaptotagmin IIa |
| chr15_+_16897554 | 6.38 |
ENSDART00000154679
|
ypel2b
|
yippee-like 2b |
| chr15_+_29085955 | 5.99 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr19_+_27479563 | 5.64 |
ENSDART00000049368
ENSDART00000185426 |
atat1
|
alpha tubulin acetyltransferase 1 |
| chr17_-_33716688 | 5.63 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr21_+_22124736 | 5.43 |
ENSDART00000130179
ENSDART00000172573 |
cul5b
|
cullin 5b |
| chr19_-_47527093 | 5.41 |
ENSDART00000171379
ENSDART00000171140 ENSDART00000114886 |
scg5
|
secretogranin V |
| chr13_+_33117528 | 5.09 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr2_-_58795694 | 4.96 |
ENSDART00000176425
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr10_+_37145007 | 4.95 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr23_+_45538932 | 4.71 |
ENSDART00000135602
|
si:ch73-290k24.6
|
si:ch73-290k24.6 |
| chr13_-_29406534 | 4.60 |
ENSDART00000100877
|
zgc:153142
|
zgc:153142 |
| chr3_-_5664123 | 4.52 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr18_+_2168695 | 4.45 |
ENSDART00000165855
|
dnaaf4
|
dynein axonemal assembly factor 4 |
| chr22_-_25033105 | 4.44 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr17_-_18888959 | 4.40 |
ENSDART00000080029
|
ak7b
|
adenylate kinase 7b |
| chr24_+_22021675 | 4.26 |
ENSDART00000081234
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr11_-_30352333 | 4.26 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr17_+_33296436 | 4.13 |
ENSDART00000123114
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr1_-_45633955 | 4.05 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr5_-_57920600 | 4.05 |
ENSDART00000132376
|
si:dkey-27p23.3
|
si:dkey-27p23.3 |
| chr10_-_17231917 | 4.03 |
ENSDART00000038577
|
rab36
|
RAB36, member RAS oncogene family |
| chr6_+_33537267 | 4.01 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr18_+_29950233 | 4.00 |
ENSDART00000146431
|
atmin
|
ATM interactor |
| chr22_+_25049563 | 3.93 |
ENSDART00000078173
|
dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr19_+_27479838 | 3.93 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr12_+_13404784 | 3.93 |
ENSDART00000167977
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr20_-_12642685 | 3.81 |
ENSDART00000173257
|
si:dkey-97l20.6
|
si:dkey-97l20.6 |
| chr25_+_5755314 | 3.70 |
ENSDART00000172088
ENSDART00000164208 |
appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr10_-_17232372 | 3.65 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr22_+_438714 | 3.61 |
ENSDART00000136491
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr17_-_22067451 | 3.59 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr21_+_21791343 | 3.56 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr19_+_43123331 | 3.54 |
ENSDART00000187836
|
CABZ01101996.1
|
|
| chr20_-_4766645 | 3.51 |
ENSDART00000147071
ENSDART00000152398 |
ak7a
|
adenylate kinase 7a |
| chr21_-_12036134 | 3.48 |
ENSDART00000031658
|
tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr18_+_5618368 | 3.43 |
ENSDART00000159945
|
ulk3
|
unc-51 like kinase 3 |
| chr25_+_21180695 | 3.42 |
ENSDART00000012923
|
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr6_+_515181 | 3.40 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr12_+_7445595 | 3.28 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr5_-_21044693 | 3.27 |
ENSDART00000140298
|
si:dkey-13n15.2
|
si:dkey-13n15.2 |
| chr2_+_59527149 | 3.26 |
ENSDART00000168568
|
CU861477.1
|
|
| chr11_+_37137196 | 3.25 |
ENSDART00000172950
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr17_-_24684687 | 3.24 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr6_-_18250857 | 3.18 |
ENSDART00000159486
|
anapc11
|
APC11 anaphase promoting complex subunit 11 homolog (yeast) |
| chr3_+_28831622 | 3.17 |
ENSDART00000184130
ENSDART00000191294 |
flr
|
fleer |
| chr6_+_18251140 | 3.15 |
ENSDART00000169752
|
ccdc40
|
coiled-coil domain containing 40 |
| chr3_+_28831450 | 3.14 |
ENSDART00000055422
|
flr
|
fleer |
| chr9_+_2452672 | 3.12 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr20_+_591505 | 3.08 |
ENSDART00000046438
|
kcnk2b
|
potassium channel, subfamily K, member 2b |
| chr17_-_8173380 | 3.05 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr24_+_38155830 | 3.01 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr17_+_33296852 | 2.98 |
ENSDART00000154580
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr20_+_33519435 | 2.95 |
ENSDART00000061829
|
drc1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr6_+_52350443 | 2.88 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr17_+_11500628 | 2.87 |
ENSDART00000155660
|
efcab2
|
EF-hand calcium binding domain 2 |
| chr10_+_15255198 | 2.82 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr20_+_52554352 | 2.81 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr10_+_15255012 | 2.81 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr7_+_38395197 | 2.77 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr1_-_21538329 | 2.75 |
ENSDART00000182798
ENSDART00000193518 ENSDART00000183873 ENSDART00000183376 |
wdr19
|
WD repeat domain 19 |
| chr14_-_52542928 | 2.68 |
ENSDART00000075749
|
ppp2r2ba
|
protein phosphatase 2, regulatory subunit B, beta a |
| chr2_-_1569250 | 2.67 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr1_-_21538673 | 2.67 |
ENSDART00000131257
|
wdr19
|
WD repeat domain 19 |
| chr2_+_33796207 | 2.67 |
ENSDART00000124647
|
kiss1ra
|
KISS1 receptor a |
| chr2_-_54039293 | 2.57 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr12_+_17620067 | 2.55 |
ENSDART00000073599
|
rsph10b
|
radial spoke head 10 homolog B |
| chr6_-_19683406 | 2.54 |
ENSDART00000158041
|
cfap52
|
cilia and flagella associated protein 52 |
| chr17_+_27545183 | 2.54 |
ENSDART00000129392
|
pacrg
|
PARK2 co-regulated |
| chr7_-_40738774 | 2.51 |
ENSDART00000084179
|
rnf32
|
ring finger protein 32 |
| chr19_+_8985230 | 2.50 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr24_+_22022109 | 2.49 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr15_+_34062460 | 2.44 |
ENSDART00000164654
|
si:dkey-30e9.6
|
si:dkey-30e9.6 |
| chr19_+_22062202 | 2.42 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr24_-_6647275 | 2.41 |
ENSDART00000161494
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr7_-_72423666 | 2.41 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr19_-_10648523 | 2.36 |
ENSDART00000059358
|
tekt2
|
tektin 2 (testicular) |
| chr11_+_43401201 | 2.32 |
ENSDART00000190128
|
vipb
|
vasoactive intestinal peptide b |
| chr13_-_36703164 | 2.32 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr17_-_2721336 | 2.29 |
ENSDART00000109582
ENSDART00000192691 ENSDART00000189381 |
spata7
|
spermatogenesis associated 7 |
| chr19_+_19976990 | 2.26 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr19_+_9091673 | 2.25 |
ENSDART00000052898
|
si:ch211-81a5.5
|
si:ch211-81a5.5 |
| chr14_-_38827442 | 2.24 |
ENSDART00000160000
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr25_-_16755340 | 2.23 |
ENSDART00000124729
ENSDART00000110859 |
ribc2
|
RIB43A domain with coiled-coils 2 |
| chr9_-_23156908 | 2.22 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr4_-_1497384 | 2.17 |
ENSDART00000093236
|
zmp:0000000711
|
zmp:0000000711 |
| chr17_+_21818093 | 2.16 |
ENSDART00000125335
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr9_+_27339505 | 2.15 |
ENSDART00000011822
|
slc10a2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr9_+_52411530 | 2.14 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr20_-_32007209 | 2.13 |
ENSDART00000021575
|
adgb
|
androglobin |
| chr11_-_12417952 | 2.13 |
ENSDART00000135694
|
si:dkey-27d5.9
|
si:dkey-27d5.9 |
| chr13_+_31205439 | 2.12 |
ENSDART00000132326
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr4_+_77919987 | 2.12 |
ENSDART00000150171
|
ttll1
|
tubulin tyrosine ligase-like family, member 1 |
| chr21_-_45349548 | 2.12 |
ENSDART00000008454
|
skp1
|
S-phase kinase-associated protein 1 |
| chr11_+_28476298 | 2.09 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr17_+_31592191 | 2.09 |
ENSDART00000153765
|
si:dkey-13p1.3
|
si:dkey-13p1.3 |
| chr14_+_24283915 | 2.08 |
ENSDART00000172868
|
klhl3
|
kelch-like family member 3 |
| chr12_+_2381213 | 2.07 |
ENSDART00000188007
|
LO018238.1
|
|
| chr3_+_17456428 | 2.05 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr3_-_12026741 | 2.05 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr16_-_24561354 | 2.05 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr25_-_13659249 | 2.04 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr14_-_38826739 | 2.03 |
ENSDART00000187633
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr15_+_31735931 | 1.98 |
ENSDART00000185681
ENSDART00000149137 |
rxfp2b
|
relaxin/insulin-like family peptide receptor 2b |
| chr20_+_13969414 | 1.97 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr3_+_24458899 | 1.96 |
ENSDART00000156655
|
cbx6b
|
chromobox homolog 6b |
| chr1_-_17675037 | 1.96 |
ENSDART00000142689
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr12_+_46582168 | 1.96 |
ENSDART00000189402
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr11_-_12512122 | 1.94 |
ENSDART00000145338
|
si:dkey-27d5.3
|
si:dkey-27d5.3 |
| chr18_-_44359726 | 1.92 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr11_-_28911172 | 1.90 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr19_+_27589201 | 1.90 |
ENSDART00000182060
|
si:dkeyp-46h3.1
|
si:dkeyp-46h3.1 |
| chr2_+_55914699 | 1.89 |
ENSDART00000157905
|
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr21_+_26726936 | 1.86 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr2_+_30786773 | 1.85 |
ENSDART00000019029
ENSDART00000145681 |
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr4_+_9609905 | 1.82 |
ENSDART00000142284
ENSDART00000150687 |
meig1
|
meiosis/spermiogenesis associated 1 |
| chr5_-_43071058 | 1.80 |
ENSDART00000165546
|
si:dkey-245n4.2
|
si:dkey-245n4.2 |
| chr4_+_34343524 | 1.79 |
ENSDART00000171694
|
si:ch211-246b8.5
|
si:ch211-246b8.5 |
| chr8_+_16990120 | 1.79 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr11_-_12379541 | 1.79 |
ENSDART00000171717
|
zgc:174353
|
zgc:174353 |
| chr11_-_12364122 | 1.78 |
ENSDART00000170129
|
zgc:174353
|
zgc:174353 |
| chr6_-_16406210 | 1.78 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr20_-_44576949 | 1.76 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr17_+_33767890 | 1.75 |
ENSDART00000193177
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr14_-_30724165 | 1.74 |
ENSDART00000020936
|
fibpa
|
fibroblast growth factor (acidic) intracellular binding protein a |
| chr2_+_45362016 | 1.71 |
ENSDART00000139154
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr13_-_6218248 | 1.69 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr8_+_31016180 | 1.69 |
ENSDART00000130870
ENSDART00000143604 |
odf2b
|
outer dense fiber of sperm tails 2b |
| chr13_-_48764180 | 1.68 |
ENSDART00000167157
|
si:ch1073-266p11.2
|
si:ch1073-266p11.2 |
| chr23_+_12134839 | 1.67 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr24_+_25467465 | 1.66 |
ENSDART00000189933
|
smpx
|
small muscle protein, X-linked |
| chr17_+_21817859 | 1.65 |
ENSDART00000143832
ENSDART00000141462 |
ikzf5
|
IKAROS family zinc finger 5 |
| chr1_+_38818268 | 1.64 |
ENSDART00000166864
|
spcs3
|
signal peptidase complex subunit 3 |
| chr8_-_14080534 | 1.64 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr14_+_30413312 | 1.62 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr21_-_45348983 | 1.61 |
ENSDART00000188017
|
skp1
|
S-phase kinase-associated protein 1 |
| chr23_-_45539306 | 1.60 |
ENSDART00000161913
|
CU681841.1
|
|
| chr23_+_7710447 | 1.60 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr11_-_12232769 | 1.59 |
ENSDART00000180116
|
si:ch211-156l18.4
|
si:ch211-156l18.4 |
| chr11_-_12232929 | 1.55 |
ENSDART00000127611
|
si:ch211-156l18.4
|
si:ch211-156l18.4 |
| chr11_+_11374021 | 1.51 |
ENSDART00000170806
|
si:dkey-23f9.8
|
si:dkey-23f9.8 |
| chr14_+_23874062 | 1.49 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr13_+_24842857 | 1.48 |
ENSDART00000123866
|
dusp13a
|
dual specificity phosphatase 13a |
| chr7_-_41881177 | 1.47 |
ENSDART00000174258
ENSDART00000018972 |
zgc:92818
|
zgc:92818 |
| chr5_+_32260502 | 1.47 |
ENSDART00000149020
|
si:ch211-158m24.12
|
si:ch211-158m24.12 |
| chr14_+_24840669 | 1.47 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr9_-_22963897 | 1.46 |
ENSDART00000133676
|
si:dkey-91i10.2
|
si:dkey-91i10.2 |
| chr5_+_24882633 | 1.46 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr6_+_6802142 | 1.45 |
ENSDART00000065566
|
dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr18_+_27101096 | 1.45 |
ENSDART00000171128
ENSDART00000086094 |
plekha7a
|
pleckstrin homology domain containing, family A member 7a |
| chr3_+_60716904 | 1.45 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr4_-_4261673 | 1.44 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr11_+_1533097 | 1.43 |
ENSDART00000066191
|
ift52
|
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr13_+_24717880 | 1.41 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr24_-_36910224 | 1.41 |
ENSDART00000079233
|
dnaaf3l
|
dynein, axonemal, assembly factor 3 like |
| chr13_-_4303641 | 1.40 |
ENSDART00000136262
|
ppp2r5d
|
protein phosphatase 2, regulatory subunit B', delta |
| chr5_-_52813442 | 1.38 |
ENSDART00000169305
|
zgc:158260
|
zgc:158260 |
| chr12_+_28955766 | 1.38 |
ENSDART00000123417
ENSDART00000139347 |
znf668
|
zinc finger protein 668 |
| chr24_+_26345427 | 1.34 |
ENSDART00000089756
|
lrrc34
|
leucine rich repeat containing 34 |
| chr9_+_48415043 | 1.33 |
ENSDART00000159930
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr15_-_1534232 | 1.30 |
ENSDART00000056763
ENSDART00000133943 |
ift80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr16_-_50203058 | 1.29 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr16_-_28709874 | 1.29 |
ENSDART00000127753
|
adam15
|
ADAM metallopeptidase domain 15 |
| chr11_-_34720629 | 1.29 |
ENSDART00000162066
ENSDART00000161471 |
dnah1
|
dynein, axonemal, heavy chain 1 |
| chr4_-_73520581 | 1.27 |
ENSDART00000171513
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr5_-_26566435 | 1.27 |
ENSDART00000146070
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr22_-_11438627 | 1.26 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr6_+_6802582 | 1.20 |
ENSDART00000189422
|
dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr18_+_50890749 | 1.19 |
ENSDART00000174109
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr13_+_24679674 | 1.18 |
ENSDART00000033090
ENSDART00000139854 |
zgc:66426
|
zgc:66426 |
| chr2_+_30787128 | 1.18 |
ENSDART00000189233
|
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr11_-_12717004 | 1.17 |
ENSDART00000122020
|
si:ch211-209f22.3
|
si:ch211-209f22.3 |
| chr13_-_8279533 | 1.16 |
ENSDART00000109666
|
si:ch211-250c4.5
|
si:ch211-250c4.5 |
| chr24_+_26345609 | 1.15 |
ENSDART00000186844
|
lrrc34
|
leucine rich repeat containing 34 |
| chr13_+_159504 | 1.13 |
ENSDART00000057248
|
ewsr1b
|
EWS RNA-binding protein 1b |
| chr9_+_48219111 | 1.13 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr4_-_73727926 | 1.13 |
ENSDART00000171414
|
si:dkey-262g12.14
|
si:dkey-262g12.14 |
| chr11_-_12437234 | 1.12 |
ENSDART00000169324
|
zgc:174355
|
zgc:174355 |
| chr20_-_28642061 | 1.11 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr7_+_7630409 | 1.08 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr19_+_5480327 | 1.06 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr1_-_6225285 | 1.05 |
ENSDART00000141653
|
mdh1b
|
malate dehydrogenase 1B, NAD (soluble) |
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx7a+rfx7b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 1.5 | 4.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.8 | 6.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.8 | 6.7 | GO:0036372 | opsin transport(GO:0036372) |
| 0.8 | 3.1 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.8 | 2.3 | GO:0060986 | regulation of endocrine process(GO:0044060) endocrine hormone secretion(GO:0060986) |
| 0.7 | 2.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.7 | 28.8 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.6 | 3.8 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.6 | 4.4 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.5 | 17.2 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.5 | 5.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.5 | 1.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.5 | 4.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.5 | 5.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.5 | 7.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.4 | 3.0 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.4 | 4.6 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.4 | 5.0 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.3 | 6.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 2.7 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.3 | 1.3 | GO:0070293 | renal absorption(GO:0070293) |
| 0.3 | 3.4 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.3 | 4.0 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.3 | 9.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.3 | 3.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.3 | 2.4 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.3 | 2.3 | GO:0043584 | nose development(GO:0043584) |
| 0.3 | 3.6 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.3 | 7.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.3 | 3.2 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.3 | 1.5 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.7 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 1.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 0.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 2.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 3.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 1.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 1.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 3.2 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 11.3 | GO:0003341 | cilium movement(GO:0003341) |
| 0.2 | 1.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 1.9 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 3.5 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.2 | 2.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 7.7 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.2 | 3.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 1.7 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 2.0 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 1.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.7 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 1.7 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 3.0 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 3.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.8 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 8.3 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.1 | 1.8 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.9 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 8.8 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.1 | 1.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.7 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 10.1 | GO:0006475 | internal protein amino acid acetylation(GO:0006475) internal peptidyl-lysine acetylation(GO:0018393) |
| 0.1 | 6.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 4.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 1.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 2.2 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.7 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.1 | 1.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.4 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 2.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 4.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 2.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 19.3 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.1 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 1.3 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 1.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 2.1 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.5 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 1.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 4.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.8 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.8 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 2.7 | GO:0006399 | tRNA metabolic process(GO:0006399) |
| 0.0 | 2.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.5 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.9 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.3 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.3 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.3 | GO:0060042 | retina morphogenesis in camera-type eye(GO:0060042) |
| 0.0 | 2.2 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 1.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.3 | 6.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.3 | 6.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.7 | 50.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.7 | 5.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.6 | 6.5 | GO:0030990 | intraciliary transport particle(GO:0030990) intraciliary transport particle B(GO:0030992) |
| 0.4 | 3.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.4 | 21.4 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.4 | 1.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.4 | 2.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 5.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.3 | 4.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 1.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.2 | 7.5 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.2 | 0.8 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.2 | 1.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 7.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.2 | 2.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 9.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 1.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 5.6 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.2 | 3.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 8.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.2 | 3.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.2 | 2.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 3.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 4.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 2.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 2.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 9.7 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 2.9 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 3.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 1.6 | 7.9 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 1.4 | 5.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.9 | 2.7 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.7 | 4.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.5 | 1.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.5 | 4.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.5 | 5.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 2.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.4 | 2.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 6.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 3.6 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 18.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 6.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 3.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.3 | 4.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 1.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 3.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 3.2 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 3.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 0.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 9.6 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.2 | 1.9 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 2.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 0.5 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 2.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 6.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 8.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.7 | GO:0030507 | spectrin binding(GO:0030507) microtubule minus-end binding(GO:0051011) |
| 0.1 | 4.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 3.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 3.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 2.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.8 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 7.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.3 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.1 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 6.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 23.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 9.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 18.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 2.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 3.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 33.5 | GO:0030234 | enzyme regulator activity(GO:0030234) |
| 0.0 | 4.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 3.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 1.3 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 1.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 4.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 17.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 7.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 3.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 3.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 3.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 17.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 3.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 1.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 2.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |