Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
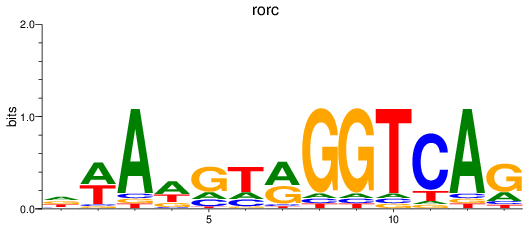
Results for rorc
Z-value: 0.62
Transcription factors associated with rorc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rorc
|
ENSDARG00000087195 | RAR-related orphan receptor C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rorc | dr11_v1_chr16_+_17913936_17913945 | 0.23 | 2.8e-02 | Click! |
Activity profile of rorc motif
Sorted Z-values of rorc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_5683147 | 9.98 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr19_+_30990129 | 6.30 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr19_+_30990815 | 6.13 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr15_-_12360409 | 5.62 |
ENSDART00000164596
|
tmprss13a
|
transmembrane protease, serine 13a |
| chr5_+_52167986 | 5.29 |
ENSDART00000162256
ENSDART00000073626 |
slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr16_+_17715243 | 4.83 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr20_-_9980318 | 4.75 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr14_+_17376940 | 3.84 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr5_+_70155935 | 3.73 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr18_+_36631923 | 3.62 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr19_+_18797623 | 3.36 |
ENSDART00000166172
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr2_-_8648440 | 3.15 |
ENSDART00000135743
|
si:ch211-71m22.3
|
si:ch211-71m22.3 |
| chr21_+_27189490 | 2.95 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr22_+_36704764 | 2.90 |
ENSDART00000146662
|
trim35-33
|
tripartite motif containing 35-33 |
| chr20_+_16750177 | 2.72 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr8_+_6576940 | 2.60 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr25_-_32888115 | 2.55 |
ENSDART00000087586
|
c2cd4a
|
C2 calcium dependent domain containing 4A |
| chr23_-_36316352 | 2.54 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr6_-_8244474 | 2.45 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr2_+_48282590 | 2.45 |
ENSDART00000035338
|
lpar5a
|
lysophosphatidic acid receptor 5a |
| chr4_+_10616626 | 2.38 |
ENSDART00000067251
ENSDART00000143690 |
cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr19_+_43314700 | 2.37 |
ENSDART00000049045
ENSDART00000133158 |
matn1
|
matrilin 1 |
| chr19_+_2590182 | 2.21 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr7_+_19835569 | 2.21 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr16_-_47426482 | 2.21 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr11_-_24458786 | 2.17 |
ENSDART00000089713
|
mxra8a
|
matrix-remodelling associated 8a |
| chr11_+_22134540 | 2.17 |
ENSDART00000190502
|
MDFI
|
si:dkey-91m3.1 |
| chr12_-_7234915 | 2.15 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr2_-_56649883 | 2.07 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr6_+_2030703 | 1.97 |
ENSDART00000109679
ENSDART00000187502 ENSDART00000191165 ENSDART00000187544 |
quo
|
quattro |
| chr3_+_59935606 | 1.89 |
ENSDART00000154157
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr2_+_58841181 | 1.85 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr2_+_27867830 | 1.76 |
ENSDART00000128193
|
rnf152
|
ring finger protein 152 |
| chr21_+_40106448 | 1.74 |
ENSDART00000100166
|
serpinf1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr17_-_31044803 | 1.67 |
ENSDART00000185941
ENSDART00000152016 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr16_+_30438041 | 1.66 |
ENSDART00000137977
|
pear1
|
platelet endothelial aggregation receptor 1 |
| chr18_-_15911394 | 1.62 |
ENSDART00000091339
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr22_-_834106 | 1.62 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr11_+_10975012 | 1.56 |
ENSDART00000192872
|
itgb6
|
integrin, beta 6 |
| chr22_+_8612462 | 1.56 |
ENSDART00000114586
|
CR450686.1
|
|
| chr10_+_44692272 | 1.54 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr11_+_10975481 | 1.49 |
ENSDART00000160488
|
itgb6
|
integrin, beta 6 |
| chr18_+_7594012 | 1.48 |
ENSDART00000062150
|
zgc:77752
|
zgc:77752 |
| chr20_+_34320635 | 1.45 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr2_-_56655769 | 1.41 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr4_-_9780931 | 1.40 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr7_-_22632518 | 1.39 |
ENSDART00000161046
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr5_-_69523816 | 1.35 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr22_-_621609 | 1.35 |
ENSDART00000137264
ENSDART00000106636 |
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr25_-_1235457 | 1.31 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr10_-_25628555 | 1.30 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr13_+_21768447 | 1.26 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr21_+_19517492 | 1.19 |
ENSDART00000123168
ENSDART00000187993 |
gzmk
|
granzyme K |
| chr16_-_27442344 | 1.18 |
ENSDART00000027545
|
erp44
|
endoplasmic reticulum protein 44 |
| chr11_-_42396302 | 1.12 |
ENSDART00000165624
|
slmapa
|
sarcolemma associated protein a |
| chr15_-_43625549 | 1.11 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr10_-_44341288 | 1.11 |
ENSDART00000166131
|
zswim6
|
zinc finger, SWIM-type containing 6 |
| chr5_+_19413838 | 1.08 |
ENSDART00000089014
ENSDART00000146215 |
myo1ha
|
myosin IHa |
| chr21_-_5881344 | 1.04 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr1_-_49498116 | 1.03 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr7_-_22632938 | 1.03 |
ENSDART00000159867
ENSDART00000165706 |
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr1_+_17527931 | 1.02 |
ENSDART00000131927
|
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr8_+_43340995 | 1.00 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr16_-_30564319 | 0.99 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr1_-_29658721 | 0.97 |
ENSDART00000132063
|
si:dkey-1h24.6
|
si:dkey-1h24.6 |
| chr5_+_19261687 | 0.91 |
ENSDART00000139401
|
atp8b5a
|
ATPase phospholipid transporting 8B5a |
| chr8_+_18463797 | 0.89 |
ENSDART00000148958
|
alkal1
|
ALK and LTK ligand 1 |
| chr6_-_49159207 | 0.88 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr2_-_38337122 | 0.87 |
ENSDART00000076523
ENSDART00000187473 |
slc7a8b
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8b |
| chr8_-_29706882 | 0.86 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr5_+_19261391 | 0.84 |
ENSDART00000089173
|
atp8b5a
|
ATPase phospholipid transporting 8B5a |
| chr25_-_17918536 | 0.83 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr3_+_32698424 | 0.83 |
ENSDART00000055340
|
fus
|
FUS RNA binding protein |
| chr8_+_18464235 | 0.83 |
ENSDART00000110571
|
alkal1
|
ALK and LTK ligand 1 |
| chr3_+_14641962 | 0.81 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr3_-_29508959 | 0.79 |
ENSDART00000055408
|
cyth4a
|
cytohesin 4a |
| chr8_+_26034623 | 0.79 |
ENSDART00000004521
ENSDART00000142555 |
arih2
|
ariadne homolog 2 (Drosophila) |
| chr19_+_42336523 | 0.77 |
ENSDART00000151304
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr20_-_30377221 | 0.76 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| chr2_-_37896965 | 0.76 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr10_-_1788376 | 0.74 |
ENSDART00000123842
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr8_-_25771474 | 0.73 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr15_-_2841677 | 0.72 |
ENSDART00000026145
ENSDART00000180290 |
AMOTL1
|
angiomotin like 1 |
| chr1_-_31140096 | 0.68 |
ENSDART00000172243
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr22_-_11124419 | 0.63 |
ENSDART00000149634
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr10_+_15454745 | 0.62 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr17_-_24364527 | 0.60 |
ENSDART00000155192
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr11_+_26363435 | 0.60 |
ENSDART00000088800
|
ppcs
|
phosphopantothenoylcysteine synthetase |
| chr10_+_575929 | 0.57 |
ENSDART00000129856
|
smad4a
|
SMAD family member 4a |
| chr8_+_25351863 | 0.56 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr9_-_52598343 | 0.55 |
ENSDART00000167922
|
xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 |
| chr7_+_48675347 | 0.55 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr2_+_49864219 | 0.53 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr7_+_24573721 | 0.53 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr1_+_15204633 | 0.53 |
ENSDART00000188410
|
itln1
|
intelectin 1 |
| chr1_-_12393060 | 0.49 |
ENSDART00000079782
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr24_+_24809877 | 0.49 |
ENSDART00000185117
|
dnajc5b
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr18_+_20225961 | 0.48 |
ENSDART00000045679
|
tle3a
|
transducin-like enhancer of split 3a |
| chr25_-_17918810 | 0.44 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr3_-_5555500 | 0.43 |
ENSDART00000176133
ENSDART00000057464 |
trim35-31
|
tripartite motif containing 35-31 |
| chr12_+_7399403 | 0.41 |
ENSDART00000103526
|
bicc1b
|
BicC family RNA binding protein 1b |
| chr8_+_26254903 | 0.39 |
ENSDART00000113763
|
slc26a6
|
solute carrier family 26, member 6 |
| chr3_+_60761811 | 0.38 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr16_-_24561354 | 0.36 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr17_-_2126517 | 0.36 |
ENSDART00000013093
|
zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr8_-_2602572 | 0.35 |
ENSDART00000110482
|
zdhhc12a
|
zinc finger, DHHC-type containing 12a |
| chr22_-_6420239 | 0.34 |
ENSDART00000148385
|
zgc:171699
|
zgc:171699 |
| chr22_-_18546241 | 0.34 |
ENSDART00000105404
ENSDART00000105405 |
cirbpb
|
cold inducible RNA binding protein b |
| chr10_-_39321367 | 0.33 |
ENSDART00000129647
|
smtlb
|
somatolactin beta |
| chr16_+_11242443 | 0.33 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr23_-_36724575 | 0.32 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr14_-_29859067 | 0.29 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr25_+_3035384 | 0.29 |
ENSDART00000184219
ENSDART00000149360 |
mpi
|
mannose phosphate isomerase |
| chr19_+_4990496 | 0.27 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr2_-_25120197 | 0.27 |
ENSDART00000132549
|
pccb
|
propionyl CoA carboxylase, beta polypeptide |
| chr9_+_13682133 | 0.23 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr6_-_43005901 | 0.20 |
ENSDART00000161071
|
si:ch73-361p23.3
|
si:ch73-361p23.3 |
| chr14_-_34700633 | 0.20 |
ENSDART00000150358
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr7_-_30227878 | 0.20 |
ENSDART00000173514
|
znf710b
|
zinc finger protein 710b |
| chr25_-_37227491 | 0.20 |
ENSDART00000156647
|
glg1a
|
golgi glycoprotein 1a |
| chr21_-_25613249 | 0.19 |
ENSDART00000137896
|
fibpb
|
fibroblast growth factor (acidic) intracellular binding protein b |
| chr25_+_11389220 | 0.13 |
ENSDART00000187787
|
AGBL1
|
si:ch73-141f14.1 |
| chr14_+_22297257 | 0.12 |
ENSDART00000113676
|
atp10b
|
ATPase phospholipid transporting 10B |
| chr4_-_4129187 | 0.08 |
ENSDART00000146889
|
asb15b
|
ankyrin repeat and SOCS box containing 15b |
| chr8_+_43016714 | 0.07 |
ENSDART00000142671
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr11_-_39118882 | 0.04 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr24_-_32408404 | 0.04 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr19_+_10860485 | 0.03 |
ENSDART00000169962
|
si:ch73-347e22.4
|
si:ch73-347e22.4 |
| chr1_-_21287724 | 0.03 |
ENSDART00000193900
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr25_+_30233638 | 0.02 |
ENSDART00000192558
ENSDART00000067057 ENSDART00000147327 ENSDART00000145924 ENSDART00000148250 |
alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr5_-_7513082 | 0.01 |
ENSDART00000158913
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
Network of associatons between targets according to the STRING database.
First level regulatory network of rorc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.0 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 1.8 | 5.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.9 | 3.6 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.8 | 3.4 | GO:1904407 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.7 | 3.7 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 2.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.3 | 1.0 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.3 | 1.0 | GO:0061182 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.3 | 2.2 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.3 | 1.2 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.3 | 1.7 | GO:0070378 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.2 | 2.5 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.2 | 1.0 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.2 | 1.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 2.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.4 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 1.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.5 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 2.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 2.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 0.3 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 3.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 3.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.4 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 1.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.6 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 1.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 3.8 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 2.4 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.3 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.5 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 2.0 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 0.6 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 3.5 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 2.2 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.0 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 1.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.1 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.2 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.8 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.8 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 2.4 | GO:0051216 | cartilage development(GO:0051216) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 10.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.6 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.1 | 13.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 2.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.3 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 10.0 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.7 | 2.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.7 | 3.5 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.5 | 5.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.5 | 1.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 2.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.4 | 3.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 1.7 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.3 | 3.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 1.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.1 | 1.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.5 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 3.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.5 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 1.0 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 1.6 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 6.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 1.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 5.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.2 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 2.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 3.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 2.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.9 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 2.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |