Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
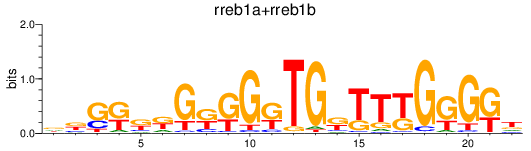
Results for rreb1a+rreb1b
Z-value: 2.60
Transcription factors associated with rreb1a+rreb1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rreb1b
|
ENSDARG00000042652 | ras responsive element binding protein 1b |
|
rreb1a
|
ENSDARG00000063701 | ras responsive element binding protein 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rreb1a | dr11_v1_chr24_-_2381143_2381143 | 0.28 | 6.0e-03 | Click! |
| rreb1b | dr11_v1_chr2_+_21000334_21000355 | 0.15 | 1.4e-01 | Click! |
Activity profile of rreb1a+rreb1b motif
Sorted Z-values of rreb1a+rreb1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_10606 | 48.09 |
ENSDART00000192650
ENSDART00000186761 |
FO704772.2
|
|
| chr5_+_817016 | 45.01 |
ENSDART00000188498
|
BX537263.3
|
|
| chr5_+_828663 | 45.00 |
ENSDART00000181776
|
BX537263.4
|
|
| chr4_-_77563411 | 33.41 |
ENSDART00000186841
|
AL935186.8
|
|
| chr21_+_28445052 | 32.59 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr4_-_77551860 | 32.45 |
ENSDART00000188176
|
AL935186.6
|
|
| chr17_-_122680 | 30.08 |
ENSDART00000066430
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr8_+_54284961 | 29.22 |
ENSDART00000122692
|
plxnd1
|
plexin D1 |
| chr12_+_13244149 | 27.12 |
ENSDART00000186984
ENSDART00000105896 |
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr4_+_90048 | 26.85 |
ENSDART00000166440
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr7_-_2039060 | 25.95 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr7_+_74150839 | 25.72 |
ENSDART00000160195
|
ppp1cbl
|
protein phosphatase 1, catalytic subunit, beta isoform, like |
| chr5_-_72125551 | 24.73 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr18_+_5547185 | 24.03 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr11_+_36243774 | 23.89 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr10_-_57270 | 22.00 |
ENSDART00000058411
|
hlcs
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
| chr7_-_73752955 | 21.77 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr18_-_50845804 | 20.16 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr8_+_554531 | 20.06 |
ENSDART00000193623
|
FO704758.2
|
|
| chr14_+_52563794 | 19.43 |
ENSDART00000168874
|
rpl26
|
ribosomal protein L26 |
| chr8_-_554540 | 19.31 |
ENSDART00000163934
|
FO704758.1
|
Danio rerio uncharacterized LOC100329294 (LOC100329294), mRNA. |
| chr13_-_51846224 | 19.10 |
ENSDART00000184663
|
LT631684.2
|
|
| chr1_+_55002583 | 18.88 |
ENSDART00000037250
|
si:ch211-196h16.12
|
si:ch211-196h16.12 |
| chr21_+_25765734 | 18.43 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr4_-_18211 | 17.82 |
ENSDART00000171737
|
ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr19_+_48117995 | 17.55 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr19_-_5380770 | 17.53 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr3_-_1190132 | 16.61 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr15_-_47479119 | 16.35 |
ENSDART00000164957
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr4_+_77948517 | 15.33 |
ENSDART00000149305
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr9_+_30294096 | 15.05 |
ENSDART00000026551
|
srpx
|
sushi-repeat containing protein, X-linked |
| chr3_-_1434135 | 14.79 |
ENSDART00000149622
|
mgp
|
matrix Gla protein |
| chr12_-_54375 | 14.67 |
ENSDART00000152304
|
si:ch1073-357b18.4
|
si:ch1073-357b18.4 |
| chr9_-_56420087 | 14.33 |
ENSDART00000112016
|
actr3
|
ARP3 actin related protein 3 homolog |
| chr1_-_59243542 | 14.17 |
ENSDART00000163021
|
mvb12a
|
multivesicular body subunit 12A |
| chr12_-_4540564 | 14.03 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr4_+_77948970 | 13.83 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr21_-_226071 | 13.81 |
ENSDART00000160667
|
nup54
|
nucleoporin 54 |
| chr13_+_22675802 | 13.57 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr17_+_53311618 | 13.44 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr17_-_53348486 | 13.34 |
ENSDART00000161183
|
CABZ01066926.1
|
|
| chr23_-_38054 | 13.11 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr8_+_3379815 | 12.99 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr1_+_27690 | 12.96 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr5_+_480119 | 12.84 |
ENSDART00000055681
|
tek
|
TEK tyrosine kinase, endothelial |
| chr3_-_1408487 | 12.69 |
ENSDART00000100845
|
bglap
|
bone gamma-carboxyglutamate (gla) protein |
| chr7_+_31838320 | 12.66 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr23_+_44580254 | 12.64 |
ENSDART00000185069
|
pfn1
|
profilin 1 |
| chr17_+_691453 | 12.29 |
ENSDART00000159271
|
fancm
|
Fanconi anemia, complementation group M |
| chr25_-_37489917 | 12.11 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr7_+_50849142 | 12.01 |
ENSDART00000073806
|
pcolceb
|
procollagen C-endopeptidase enhancer b |
| chr1_+_227241 | 12.00 |
ENSDART00000003317
|
tfdp1b
|
transcription factor Dp-1, b |
| chr4_-_68569527 | 11.94 |
ENSDART00000192091
|
BX548011.5
|
|
| chr14_+_34514336 | 11.92 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr6_-_21189295 | 11.61 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr3_-_60027255 | 11.60 |
ENSDART00000189252
ENSDART00000154684 |
recql5
|
RecQ helicase-like 5 |
| chr24_+_42131564 | 11.40 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr13_+_41022502 | 11.39 |
ENSDART00000026808
|
dkk1a
|
dickkopf WNT signaling pathway inhibitor 1a |
| chr18_-_49078428 | 11.01 |
ENSDART00000160702
ENSDART00000174103 |
BX663503.3
|
|
| chr2_+_7192966 | 10.78 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr18_+_173603 | 10.67 |
ENSDART00000185918
|
larp6a
|
La ribonucleoprotein domain family, member 6a |
| chr3_+_18398876 | 10.61 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr4_+_55758103 | 10.48 |
ENSDART00000185964
|
CT583728.23
|
|
| chr6_+_27514465 | 10.47 |
ENSDART00000128985
ENSDART00000079397 |
ryk
|
receptor-like tyrosine kinase |
| chr23_+_22873415 | 10.46 |
ENSDART00000135130
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr1_-_9104631 | 10.46 |
ENSDART00000146642
|
si:ch211-14k19.8
|
si:ch211-14k19.8 |
| chr6_+_36821621 | 10.45 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr20_+_2739804 | 10.44 |
ENSDART00000152655
|
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr1_-_48933 | 10.35 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr14_-_9522364 | 10.24 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr25_-_1124851 | 10.24 |
ENSDART00000067558
|
spg11
|
spastic paraplegia 11 |
| chr9_-_216527 | 10.12 |
ENSDART00000163068
|
aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr3_-_1400309 | 10.02 |
ENSDART00000159893
|
wbp11
|
WW domain binding protein 11 |
| chr4_-_64703 | 10.00 |
ENSDART00000167851
|
CU856344.1
|
|
| chr25_+_150570 | 9.93 |
ENSDART00000170892
|
adam10b
|
ADAM metallopeptidase domain 10b |
| chr2_-_51511494 | 9.91 |
ENSDART00000161392
ENSDART00000160877 |
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr13_-_214122 | 9.85 |
ENSDART00000169273
|
ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr8_-_28449782 | 9.80 |
ENSDART00000062702
|
cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr2_-_51700709 | 9.75 |
ENSDART00000188601
|
tgm1l1
|
transglutaminase 1 like 1 |
| chr25_-_37501371 | 9.70 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr2_+_36007449 | 9.58 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr9_+_22764235 | 9.45 |
ENSDART00000090875
|
tnfaip6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr7_-_5162292 | 9.41 |
ENSDART00000084218
|
zgc:195075
|
zgc:195075 |
| chr11_+_45233348 | 9.30 |
ENSDART00000173150
|
tmc6b
|
transmembrane channel-like 6b |
| chr24_+_38671054 | 9.20 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr15_-_44663712 | 9.15 |
ENSDART00000192124
|
CABZ01061894.1
|
|
| chr25_-_37465064 | 9.13 |
ENSDART00000186128
|
zgc:158366
|
zgc:158366 |
| chr21_-_44009169 | 9.03 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr24_-_41478917 | 8.91 |
ENSDART00000192192
|
CABZ01084131.1
|
|
| chr7_-_55454406 | 8.88 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr25_+_1732838 | 8.65 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr22_+_661505 | 8.59 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr1_+_59538755 | 8.52 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr9_-_23807032 | 8.49 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr3_+_301479 | 8.36 |
ENSDART00000165169
|
CABZ01075611.1
|
|
| chr19_+_32856907 | 8.31 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr17_+_132555 | 8.29 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr21_+_233271 | 8.26 |
ENSDART00000171440
|
dtwd2
|
DTW domain containing 2 |
| chr22_+_10090673 | 8.25 |
ENSDART00000186680
|
si:dkey-102c8.3
|
si:dkey-102c8.3 |
| chr19_-_5364649 | 8.09 |
ENSDART00000004812
|
cyt1
|
type I cytokeratin, enveloping layer |
| chr6_+_27338512 | 8.08 |
ENSDART00000155004
|
klhl30
|
kelch-like family member 30 |
| chr25_+_236753 | 8.06 |
ENSDART00000153663
|
pus7
|
pseudouridylate synthase 7 (putative) |
| chr17_+_15388479 | 8.06 |
ENSDART00000052439
|
si:ch211-266g18.6
|
si:ch211-266g18.6 |
| chr25_-_37422859 | 8.04 |
ENSDART00000156018
|
cd44b
|
CD44 molecule (Indian blood group) b |
| chr20_+_2669017 | 7.97 |
ENSDART00000058777
|
slc35a1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr17_+_53418445 | 7.88 |
ENSDART00000097631
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr17_+_53311243 | 7.86 |
ENSDART00000160241
ENSDART00000160009 ENSDART00000162239 |
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr9_+_23714406 | 7.81 |
ENSDART00000189445
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr10_+_24445698 | 7.71 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr21_+_25625026 | 7.46 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr5_+_23630384 | 7.38 |
ENSDART00000013745
|
cx39.9
|
connexin 39.9 |
| chr5_+_337215 | 7.33 |
ENSDART00000167982
|
rnf170
|
ring finger protein 170 |
| chr9_+_10692905 | 7.27 |
ENSDART00000061499
|
cxcr4b
|
chemokine (C-X-C motif), receptor 4b |
| chr17_-_5860222 | 7.27 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr7_+_53199763 | 7.22 |
ENSDART00000160097
|
cdh28
|
cadherin 28 |
| chr15_+_47362728 | 7.19 |
ENSDART00000180712
|
CABZ01099833.1
|
|
| chr1_+_59533317 | 7.15 |
ENSDART00000166821
|
sp6
|
Sp6 transcription factor |
| chr2_-_58581663 | 7.13 |
ENSDART00000004431
|
epb41l3b
|
erythrocyte membrane protein band 4.1-like 3b |
| chr16_-_54357200 | 6.84 |
ENSDART00000170753
|
CABZ01111454.1
|
|
| chr11_+_45461853 | 6.76 |
ENSDART00000173409
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr20_-_54377933 | 6.73 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr16_-_30885838 | 6.73 |
ENSDART00000131356
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr8_+_44475793 | 6.70 |
ENSDART00000190118
|
si:ch73-211l2.3
|
si:ch73-211l2.3 |
| chr10_-_45058886 | 6.59 |
ENSDART00000159347
|
mrps24
|
mitochondrial ribosomal protein S24 |
| chr19_+_638339 | 6.57 |
ENSDART00000148705
|
tert
|
telomerase reverse transcriptase |
| chr8_+_45294767 | 6.48 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr2_-_8648440 | 6.48 |
ENSDART00000135743
|
si:ch211-71m22.3
|
si:ch211-71m22.3 |
| chr10_+_23537576 | 6.39 |
ENSDART00000130515
|
CR847844.1
|
|
| chr14_+_48812189 | 6.26 |
ENSDART00000161454
|
mmp17b
|
matrix metallopeptidase 17b |
| chr3_+_60007703 | 6.23 |
ENSDART00000157351
ENSDART00000153928 ENSDART00000155876 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr11_+_45343245 | 6.21 |
ENSDART00000160904
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr20_-_38746889 | 6.19 |
ENSDART00000140275
|
trim54
|
tripartite motif containing 54 |
| chr2_+_47754419 | 6.17 |
ENSDART00000188221
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr19_-_81851 | 6.15 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr19_+_917852 | 6.10 |
ENSDART00000082466
|
tgfbr2a
|
transforming growth factor beta receptor 2a |
| chr23_-_9768700 | 6.08 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr25_+_4750972 | 6.04 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr12_+_2677303 | 6.03 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr15_-_34458495 | 5.99 |
ENSDART00000059954
|
meox2a
|
mesenchyme homeobox 2a |
| chr9_+_51891 | 5.97 |
ENSDART00000163529
|
zgc:158316
|
zgc:158316 |
| chr10_-_74408 | 5.95 |
ENSDART00000100073
ENSDART00000141723 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr11_-_40147032 | 5.94 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr25_+_29474583 | 5.90 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr1_-_40016058 | 5.90 |
ENSDART00000165373
|
cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr4_-_53370 | 5.88 |
ENSDART00000180254
ENSDART00000186529 |
CU856344.2
|
|
| chr23_-_969844 | 5.86 |
ENSDART00000127037
|
cdh26.2
|
cadherin 26, tandem duplicate 2 |
| chr17_+_51517750 | 5.81 |
ENSDART00000180896
ENSDART00000193528 |
pxdn
|
peroxidasin |
| chr6_+_59984772 | 5.81 |
ENSDART00000048449
|
mtrf1
|
mitochondrial translational release factor 1 |
| chr2_+_59041081 | 5.81 |
ENSDART00000067736
|
stk11
|
serine/threonine kinase 11 |
| chr10_+_22772084 | 5.77 |
ENSDART00000144845
ENSDART00000165222 |
tmem88a
|
transmembrane protein 88 a |
| chr1_-_5543065 | 5.74 |
ENSDART00000103755
|
fn1b
|
fibronectin 1b |
| chr6_+_392815 | 5.70 |
ENSDART00000163142
|
cyth4b
|
cytohesin 4b |
| chr18_-_226800 | 5.68 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr14_+_94946 | 5.62 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr6_-_43677125 | 5.62 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr25_-_10724947 | 5.57 |
ENSDART00000184490
|
lrp5
|
low density lipoprotein receptor-related protein 5 |
| chr3_-_30685401 | 5.54 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr22_+_997838 | 5.52 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr5_-_26181863 | 5.51 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr23_-_306796 | 5.51 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr6_-_442163 | 5.42 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr5_-_69621227 | 5.41 |
ENSDART00000178543
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr11_-_19501439 | 5.39 |
ENSDART00000158328
ENSDART00000103983 |
slc2a9l1
|
solute carrier family 2 (facilitated glucose transporter), member 9-like 1 |
| chr4_+_76619791 | 5.23 |
ENSDART00000184042
|
ms4a17a.8
|
membrane-spanning 4-domains, subfamily A, member 17A.8 |
| chr3_+_1223824 | 5.21 |
ENSDART00000065922
|
wbp2nl
|
WBP2 N-terminal like |
| chr23_-_46201008 | 5.18 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr1_+_176583 | 5.16 |
ENSDART00000168760
ENSDART00000160425 |
LAMP1
|
lysosomal associated membrane protein 1 |
| chr6_-_53326421 | 5.16 |
ENSDART00000191740
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr25_+_5044780 | 5.04 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr13_-_45523026 | 4.98 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr19_+_2619444 | 4.95 |
ENSDART00000169483
|
fam126a
|
family with sequence similarity 126, member A |
| chr19_-_5358443 | 4.94 |
ENSDART00000105036
|
cyt1l
|
type I cytokeratin, enveloping layer, like |
| chr4_-_170120 | 4.94 |
ENSDART00000171333
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr7_+_35229645 | 4.94 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr5_-_417495 | 4.91 |
ENSDART00000180586
ENSDART00000189408 |
HOOK3
|
hook microtubule tethering protein 3 |
| chr9_+_21281652 | 4.89 |
ENSDART00000113352
|
lats2
|
large tumor suppressor kinase 2 |
| chr9_+_56194410 | 4.87 |
ENSDART00000168530
|
LO018176.1
|
|
| chr12_+_49135755 | 4.84 |
ENSDART00000153460
|
si:zfos-911d5.4
|
si:zfos-911d5.4 |
| chr4_+_76722754 | 4.83 |
ENSDART00000153867
|
ms4a17a.3
|
membrane-spanning 4-domains, subfamily A, member 17A.3 |
| chr21_+_45510448 | 4.81 |
ENSDART00000160494
ENSDART00000167914 |
fnip1
|
folliculin interacting protein 1 |
| chr5_+_19337108 | 4.78 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr21_+_170038 | 4.77 |
ENSDART00000157614
|
klhl8
|
kelch-like family member 8 |
| chr17_-_4395373 | 4.75 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr12_+_2676878 | 4.72 |
ENSDART00000185909
|
antxr1c
|
anthrax toxin receptor 1c |
| chr18_-_45617146 | 4.72 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr12_+_23892972 | 4.71 |
ENSDART00000152852
|
svila
|
supervillin a |
| chr8_-_53488832 | 4.68 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr12_-_48312647 | 4.68 |
ENSDART00000114415
|
ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr18_-_127558 | 4.66 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr12_+_10053852 | 4.66 |
ENSDART00000078497
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr15_-_47468085 | 4.61 |
ENSDART00000164438
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr9_+_36946340 | 4.61 |
ENSDART00000135281
|
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr25_+_25464630 | 4.60 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr11_+_29537756 | 4.58 |
ENSDART00000103388
|
wu:fi42e03
|
wu:fi42e03 |
| chr5_+_71802014 | 4.53 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr7_-_74090168 | 4.44 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr5_-_6508250 | 4.36 |
ENSDART00000060535
|
crybb3
|
crystallin, beta B3 |
| chr7_+_22293894 | 4.36 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr15_-_18223769 | 4.30 |
ENSDART00000139966
|
or131-1
|
odorant receptor, family H, subfamily 131, member 1 |
| chr7_+_35229805 | 4.19 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rreb1a+rreb1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 32.6 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 7.3 | 21.8 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 6.0 | 24.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 5.3 | 53.1 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 4.7 | 14.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 4.6 | 13.8 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 4.0 | 28.2 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 3.4 | 10.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 3.0 | 11.9 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 2.5 | 12.7 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 2.2 | 6.6 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 1.9 | 5.8 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 1.9 | 21.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 1.9 | 5.8 | GO:2000726 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 1.9 | 7.6 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 1.9 | 5.6 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 1.8 | 5.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 1.8 | 8.9 | GO:0010481 | ventriculo bulbo valve development(GO:0003173) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 1.7 | 11.6 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 1.6 | 6.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 1.6 | 4.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 1.4 | 29.2 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 1.3 | 10.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 1.2 | 12.3 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 1.2 | 10.7 | GO:1901998 | toxin transport(GO:1901998) |
| 1.2 | 4.8 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 1.2 | 1.2 | GO:0010885 | regulation of cholesterol storage(GO:0010885) |
| 1.2 | 12.7 | GO:0033273 | response to vitamin(GO:0033273) |
| 1.1 | 17.9 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 1.1 | 17.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 1.1 | 8.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 1.0 | 10.5 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 1.0 | 5.2 | GO:0007343 | egg activation(GO:0007343) |
| 1.0 | 7.0 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.9 | 16.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.9 | 13.7 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.9 | 2.7 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.9 | 3.5 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.8 | 2.4 | GO:0003218 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.8 | 21.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.8 | 12.8 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.7 | 2.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.7 | 3.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.7 | 7.9 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.7 | 5.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.7 | 14.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.7 | 10.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.6 | 5.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.6 | 5.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.6 | 29.4 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.6 | 11.4 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.6 | 2.9 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.6 | 3.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.6 | 5.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.5 | 1.6 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.5 | 5.6 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.5 | 4.4 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.5 | 8.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.5 | 4.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.5 | 12.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.4 | 2.7 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.4 | 4.9 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.4 | 6.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 0.9 | GO:0090190 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.4 | 1.7 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.4 | 17.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.4 | 2.4 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.4 | 6.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.4 | 10.7 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.4 | 12.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.4 | 18.4 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.4 | 1.1 | GO:1903430 | branchiomotor neuron axon guidance(GO:0021785) negative regulation of cell maturation(GO:1903430) |
| 0.3 | 9.9 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.3 | 14.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.3 | 11.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.3 | 2.6 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 8.1 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.3 | 1.9 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 3.8 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.3 | 4.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.3 | 6.1 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.3 | 9.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.3 | 4.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.3 | 1.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.3 | 1.1 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.3 | 2.1 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.2 | 9.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.2 | 5.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 4.7 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.2 | 2.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.2 | 10.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 6.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 5.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.2 | 4.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 4.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 2.6 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 9.6 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.2 | 8.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 3.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 10.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 5.0 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.2 | 0.3 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.1 | 2.0 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 2.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 2.3 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.9 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 2.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 1.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 2.0 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 11.4 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 2.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 9.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 11.9 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.1 | 8.1 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 7.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 7.0 | GO:0016571 | histone methylation(GO:0016571) |
| 0.1 | 4.1 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 1.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 13.7 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 6.1 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 7.5 | GO:0009913 | epidermal cell differentiation(GO:0009913) |
| 0.1 | 16.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 6.2 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 3.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 9.7 | GO:0051169 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.1 | 2.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 3.7 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 0.7 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 3.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 6.9 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 | 4.0 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.1 | 22.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 2.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.7 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 3.2 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 1.4 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.1 | 2.9 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.1 | 1.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 4.5 | GO:0001503 | ossification(GO:0001503) |
| 0.1 | 10.7 | GO:0016570 | histone modification(GO:0016570) |
| 0.0 | 1.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.0 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 3.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 4.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 1.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 5.3 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 6.6 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 4.0 | GO:0045165 | cell fate commitment(GO:0045165) |
| 0.0 | 7.3 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 1.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 3.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 3.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 4.2 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 1.5 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 16.1 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 3.4 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 8.5 | GO:0051726 | regulation of cell cycle(GO:0051726) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 3.4 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.9 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 4.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.5 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 21.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 2.2 | 6.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 1.9 | 30.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.5 | 13.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 1.4 | 29.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 1.3 | 6.5 | GO:1990923 | PET complex(GO:1990923) |
| 1.3 | 16.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.2 | 53.1 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 1.2 | 3.6 | GO:0043034 | costamere(GO:0043034) |
| 1.2 | 24.7 | GO:0031430 | M band(GO:0031430) |
| 1.1 | 12.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.0 | 2.9 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 1.0 | 16.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.9 | 13.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.9 | 8.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.9 | 21.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.8 | 14.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.4 | 2.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.4 | 4.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 2.8 | GO:0035060 | brahma complex(GO:0035060) |
| 0.3 | 5.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 1.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 3.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 6.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.3 | 6.1 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.3 | 34.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.3 | 2.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 25.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 10.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 9.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 15.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.2 | 10.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 2.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 18.7 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.2 | 4.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 0.6 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 8.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 18.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 3.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 8.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.1 | 24.6 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 2.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 6.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 9.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 7.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 8.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 0.6 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 3.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 4.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 17.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 5.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 5.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 6.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 4.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 7.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 3.2 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 5.2 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 32.6 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 6.0 | 24.0 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 3.6 | 17.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 3.3 | 9.9 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 2.7 | 23.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 2.3 | 11.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 2.2 | 6.6 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 1.3 | 21.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.2 | 12.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 1.2 | 4.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 1.2 | 12.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.1 | 18.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.1 | 5.6 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 1.0 | 5.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 1.0 | 6.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 1.0 | 4.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.9 | 29.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.8 | 6.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.8 | 8.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.8 | 9.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.8 | 8.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.7 | 14.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.7 | 5.7 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.7 | 7.9 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.7 | 3.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.6 | 17.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.6 | 10.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.6 | 10.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.6 | 5.8 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.6 | 18.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 2.7 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.5 | 13.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.5 | 5.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 31.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.4 | 8.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.4 | 10.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.4 | 6.7 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.4 | 5.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.4 | 2.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 5.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.3 | 30.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.3 | 22.0 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.3 | 8.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 10.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 3.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 8.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.3 | 4.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.3 | 5.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 3.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 1.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 2.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 10.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 4.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 2.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 2.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.9 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.2 | 4.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 4.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 1.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 2.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 5.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 14.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 2.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 25.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 61.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 10.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 5.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 3.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 7.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.4 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 8.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 2.6 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 7.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 3.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 0.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 5.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 7.4 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.1 | 18.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 6.5 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 19.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 8.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 3.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 5.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 9.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 2.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 3.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 39.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 3.9 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.4 | GO:0015645 | fatty acid ligase activity(GO:0015645) |
| 0.0 | 1.3 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 42.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 1.3 | 15.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.1 | 27.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 1.0 | 28.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.5 | 12.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.5 | 9.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.4 | 9.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 30.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 11.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.4 | 6.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.3 | 5.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 2.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 39.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 12.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 3.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 4.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 5.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 3.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 3.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 20.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.6 | 29.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.8 | 22.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.7 | 10.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.6 | 23.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.6 | 12.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.5 | 3.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.5 | 12.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.4 | 3.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.4 | 5.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 5.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 12.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 15.6 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.3 | 38.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 3.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 7.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 1.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 4.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 7.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 3.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 9.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 11.4 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 1.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 10.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 4.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 2.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 5.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 4.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |